Introduction
Acute gastroenteritis (AGE) is characterized by the inflammation of the gastrointestinal tract and is commonly caused by viral, bacterial, or parasitic pathogens [1]. Deaths from acute diarrheal illness, one of the most common diseases worldwide, are gradually decreasing; however, 440,000 children under the age of 5 years still die annually from this disease, according to the 2018 World Health Organization report [2]. Since the introduction of the rotavirus vaccine, although a reduction in the incidence and mortality of diarrheal illness has been reported, it remains a major disease affecting children. In Korea, examination of the pathogens responsible for acute diarrheal diseases began in 1972, linking the Korea Centers for Disease Control and Prevention (KCDC), which was reorganized into the KDCA (Korea Disease Control and Prevention Agency), Public Health Center, and Institute of Health and Environment. The monitoring laboratory for acute diarrheal disease (EnterNet-Korea) started operations in 2003. Since 2005, the KCDC has been monitoring pathogens that cause AGE, in patients who visit hospitals with the major symptoms of diarrhea and abdominal pain through a waterborne food disease monitoring network operation project involving 70–192 medical institutions, although the number of organizations participating each year was different. Rotavirus, norovirus, enterovirus, and astrovirus were initially monitored, but sapovirus was added to the watch list in 2009 [3].
The results and related data of the aforementioned projects were first provided as “Infectious Disease Control Information” in February 1990 and then as the “KCDC Public Health Weekly Report” starting in April 2008 and continuing weekly thereafter through the KCDC website (www.cdc.go.kr) and related academic societies and organizations [4-7]. This gradually helped establish a foundation for comprehensive information, prevention, and proactive responses to the detection of pathogens that cause diarrhea by analyzing information such as weekly detection rates, trends in separation rates over the past 4 weeks, and separation rates from the previous year through the monitoring of waterborne food diseases on a nationwide scale [8].
Herein, KCDC data and articles in the PubMed/MEDLINE and KoreaMed databases (www.koreamed.org) were mainly compared and analyzed for AGE patients less than 18 years. This review aimed to help identify the characteristics of the causative pathogens of AGE, assess their prevalence over time, and predict the future patterns of prevalence, preparedness, and treatment of diarrheal illness caused by each pathogen.
Viral gastroenteritis
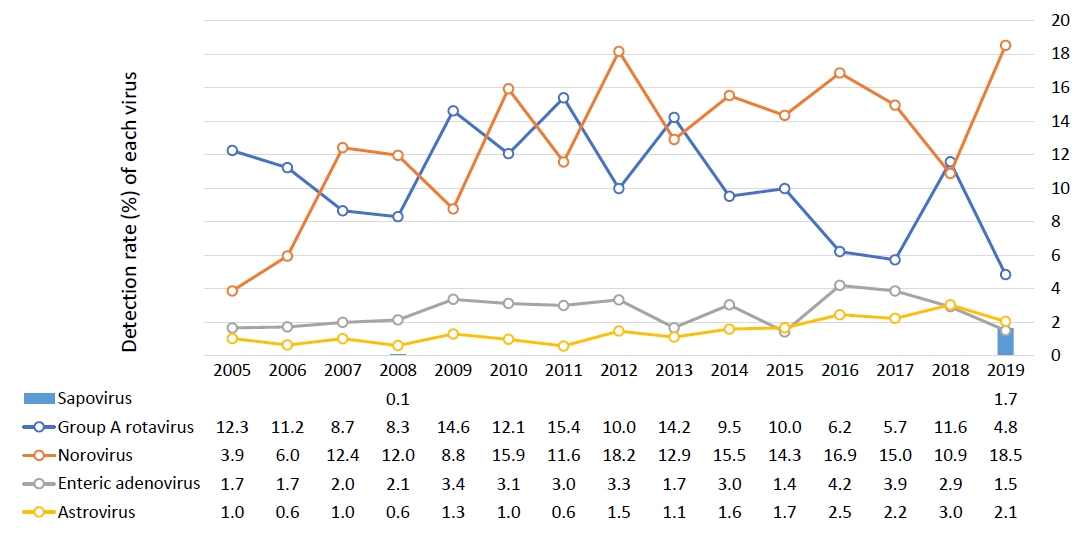
It is difficult to distinguish between viral and bacterial gastroenteritis by clinical symptoms or general laboratory tests alone; rather, specific diagnostic tests are needed for confirmation [9]. According to a survey conducted by the KCDC since 2005, children under 5 years of age have a continuous viral detection rate of around 30%, while those aged 5 years or older have a detection rate of around 10%–20%. Until 2006, the most common cause of viral gastroenteritis in children under the age of 5 years was group A rotavirus, but its incidence has gradually decreased since the introduction of the rotavirus vaccine; thus, since 2012, norovirus has been consistently reported as the most common causative virus (Fig. 1) [8,10-24].
The detection rates of viruses from 2008–2015 showed that norovirus and rotavirus changed every 2 years, apparently due to a biannual mutation, especially for norovirus, which was supported by the detection of the 2012 GII.4 Sydney and 2014 GII.17 variants [7]. In a 2013 single-hospital study of only infants, viruses accounted for less than 50% of the cases caused by bacteria, with norovirus, rotavirus, and adenovirus being the main causes [25].
Kim et al. [26] also identified that norovirus was the most common virus in children hospitalized with acute AGE, while rotavirus infection was significantly more frequent in children older than 24 months of age and more common in older age groups than other enteric viruses. The common viruses were similar to the KCDC data in the order of norovirus, rotavirus, enteric adenovirus, and astrovirus in tertiary hospital studies [27,28].
Between 2013 and 2019, acute diarrhea-causing viruses in Korea were detected most commonly among those under 5 years of age, and the detection rate was high until April of the following year, beginning to increase in October. The results for 2019 showed that norovirus and rotavirus were most frequently detected between November and June of the following year, and there were many cases of astrovirus and sapovirus in June and July. The most common viruses were norovirus (55%) and Group A rotavirus (28%), while adenovirus, astrovirus, and sapovirus accounted for less than 10% of cases, showing similar results over the previous 2 years [10,12].
1. Norovirus
The genus Norovirus belongs to the family Caliciviridae. Norovirus has a short incubation period of 12–48 hours and can infect any age group, and types I and II of the 5 genotypes are mainly pathogenic [29]. The norovirus RNA genome contains 3 open reading frames, one of which encodes polyprotein, which is differentiated into a nonstructural protein. VP1, the main capsid protein with 2 domains, forms a virus with minor capsid protein VP2. The reflector binding site appears to be in the P domain and is genetically classified according to VP1 and NS7. Human noroviruses are classified into 3 genogroups and various genotypes and subclassified according to the new variants. The most common genotype, GII.4, causes pandemics via antigenic drift and has emerged every 2–3 years; more recently, a trend by GII.17 has emerged [9,30].
The rate of infection increased after a mass outbreak of food poisoning related to school meals in 2006 that was linked to ground water contamination with norovirus. In 2007, more than half of the acute AGE viral infections were found to be caused by norovirus. Since mid-November 2010, the detection rate of norovirus has more than doubled from that of the previous year to over 30% [31-35].
The annual changes in norovirus genotypes identified in Korea are as follows: between 2004 and 2011, several GI (GI-1, GI-2, GI-3, GI-4, GI-5, GI-6, GI-7, GI-8, GI-9, GI-10, GI-12, and GI-13) and GII genotypes (GII-1, GII-2, GII-3, GII-4, GII-6, GII-7, GII-8, GII-12, GII-13, GII-14, GII-15, GII-16, and GII-17) were detected [11,31,36-38], and some variants such as GII-4/2006b, GII-4/GII-3, and GII-6/GII-14 were also detected in children [39-42].
2. Rotavirus
Of the 10 types that can infect humans, including tentative group A–I and tentative J types, group A is clinically the most important. The rotavirus genome encodes 6 structural viral proteins (VP1, VP2, VP3, VP4, VP6, and VP7) and 5 to 6 nonstructural proteins (NSP1, NSP2, NSP3, NSP4, NSP5, and NSP6) that are located on the virus surface and function as cell attachment proteins and elicit neutralized antibodies independently. Although rotavirus is genetically classified as GP7 (as G type) and VP4 (as P type), rotavirus-type specificities exist in various combinations of 14 G and 17 P types in humans and only 6 combinations (G1P[8], G2P[4], G3P[8], G4P[8], G9P[8], and G12P[8]) are clinically important [9,44].
After the introduction of the rotavirus vaccine, common genotypes of rotavirus (G1P[8], G3P[8], G4P[6], G2P[4], G9P[8], G1P[6], G3P[6], and G1P[4]P[8]) and mixed combinations were identified between 2008 and 2015 [34,35,45-47]. Rotavirus enteritis was also identified in children with rotavirus vaccination, but less commonly than that in the unvaccinated group (28% vs. 40%) in a multicenter study [47]. In neonates, G4P[6] was the most predominant type regardless of preterm birth [48]. In 2019, G8P[8], G9P[8], and G2P[4] were the 3 major genotypes. Seasonally, rotavirus used to be frequently detected twice a year, between March and April and October and December, but recently, it has appeared most frequently in February and March [10,12,43]. The effectiveness of vaccination is estimated to be about 80% [47,49].
3. Human adenovirus
All adenovirus genotypes that cause gastrointestinal diseases, including subgenus F (Ad40, Ad41), reportedly have a detection rate of 1.7%–4.2%, and studies conducted from 2014 to 2017 showed that type 41 species F was the most prevalent genotype (61%), followed by HAdV-2 of species C (14%). Other genotypes, including HAdV-3, HAdV-1, HAdV-5, HAdV-6, HAdV-31, HAdV-40, HAdV-12, HAdV-7, HAdV-4, HAdV-37, and HAdV-55, have also been identified [10,50,51].
4. Human astrovirus
Classic human astrovirus, which has 8 serotypes (genotypes), accounts for 2%–9% of childhood nonbacterial AGE cases worldwide [52].
5. Other viruses
Sapovirus infection is common in young children, mostly under 5 years of age. Sapovirus infection is milder than that caused by norovirus and mostly does not lead to the onset of symptoms [56]. Although sapovirus has a detection rate of less than 1%, it has been increasing recently in Korea [7]. Between 2013 and 2015, the main genotype was identified as the GI group, followed by the GII and GV groups [10]; however, GV has not been detected since 2016 [12].
Human bocavirus can cause gastroenteritis in children, and genotype 1 is the most predominant in Korea [57,58].
Salivirus, also known as klassevirus, is a member of the family Picornaviridae and was first identified in 2009 in Korea in the feces of children with AGE. The association between AGE and this virus is thought to occur through the low rate of coinfection with other viral agents and non-detection of the virus in nasopharyngeal aspirates [59].
Bacterial gastroenteritis
Since 2007, we have focused on the statistics of major bacterial pathogens published by the KCDC as well as the published findings of domestic and international studies. KCDC surveys identified 10 major bacterial pathogens, namely, pathogenic Escherichia coli, Salmonella spp., Shigella spp., Vibrio parahaemolyticus, Staphylococcus aureus, Clostridium perfringens, Bacillus cereus, Listeria monocytogenes, Yersinia enterocolitica, and Campylobacter jejuni, and the test was conducted on major serum and toxin types in each bacterium known to cause diarrhea [13,60].
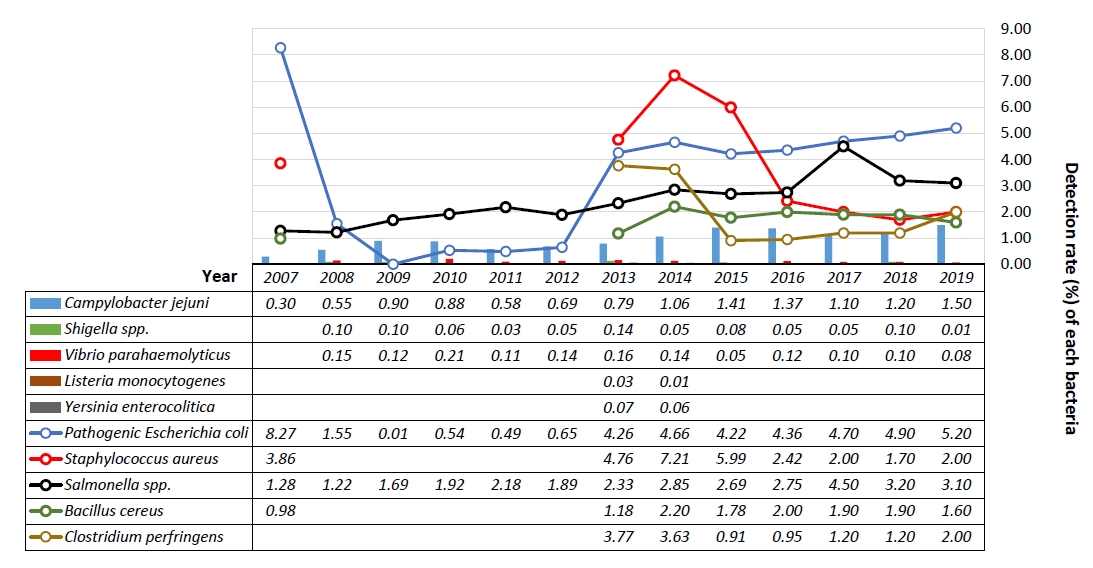
From 2004 to 2019, the bacterial detection rate was 3%–20%, which was generally high between June and September, but the distribution has been found to be widening. Particularly, the gram-positive bacteria, S. aureus and C. perfringens, are evenly distributed throughout the year. It also has the highest detection rate of around 50% among all age groups [61].
Since 2015, pathogenic E. coli has been increasingly detected and is the most common, followed by Salmonella spp., S. aureus, B. cereus, C. jejuni, C. perfringens, and Shigella spp., while L. monocytogenes, V. parahaemolyticus, and V. parahaemolyticus are relatively rare. Based on the detection rate of individual bacteria among the total bacteria detected, the detection rate of Salmonella spp., C. jejuni, and C. perfringens is gradually increasing, whereas that of B. cereus and S. aureus is relatively decreasing. C. jejuni is an important causative pathogen in food poisoning, and its detection rate has been increasing in recent years, although in Korea, the rate is not higher than that of pathogenic E. coli and Salmonella spp. Chicken has been identified as a major source of infection for C. jejuni worldwide (Fig. 2) [5,6,10,13-22,62].
1. S. aureus
Staphylococcus is the most common cause of food poisoning worldwide, with 2–8 hours of rapid manifestation caused mainly by staphylococcal enterotoxins A and B [63]. S. aureus was the most common cause of childhood bacterial diarrhea in a pre-2010 Seoul area study, affecting about 50% of the cases, and the most common of all bacteria in recent single-organ studies except for redundant infections. Causative bacteria have been identified in the following order: pathogenic E. coli, Salmonella spp., Campylobacter spp., C. perfringens, B. cereus, and Shigella spp. In older children, Campylobacter spp. was the most common [64,65]. In a domestic pediatric staphylococcal food poisoning study, seg (83%) and sei (77%) were the most common enterotoxin genes, whereas sea (28%) was relatively uncommon, unlike previous studies (64). S. aureus and C. perfringens were the most common bacteria in an infant study [25].
2. Enteric E. coli
Between 2015 and 2016, the distribution of E. coli showed that enteropathogenic E. coli (EPEC) was the most common, followed by enterotoxigenic E. coli (ETEC) and enterohemorrhagic E. coli (EHEC), and there were no cases of enteroinvasive E. coli (EIEC). Between 2013 and 2016, a single-hospital study found that E. coli accounted for about 15% of all pediatric cases of acute enteritis; EPEC (54%) was the most frequent E. coli pathotype, followed by EAEC, ETEC, and Shiga toxin-producing E. coli [66].
3. Nontyphoidal salmonellosis
The detection rate of Salmonella enterica serotype Virchow has increased rapidly due to outbreaks, and the detection rate of S. Enteritis (48%) and S. enterica Typhimurium continue to remain high. The most common serovar has changed from S. Typhimurium to S. Enteritidis; since 2013, the separation rate of Salmonella I 4,[5],12:i:- has markedly increased, and various serovars such as S. Newport, S. Agona, S. Risen, S. Panama, S. Barely, S. infantis, S. Virchow, S. Thompson, and S. Montevideo have started appearing. In 2015, the serovars were identified in order of S. Enteritidis (24%), Salmonella I 4,[5],12:i:- (13%), S. Bareilly (11%), and S. Typhimurium (9%) [67]. Between 2003 and 2012, a single-hospital study of children found that serogroup D was the most common (60%), followed by serogroups B, C, and E. The monthly incidence of nontyphoidal salmonellosis was higher from May to October than from November to April [68].
4. Campylobacter jejuni
C. jejuni infection involving chicken or seafood and the rare C. coli infection involving swine are known to be indistinguishable based on clinical manifestation alone [69]. C. jejuni infections have nearly doubled since investigations began in Korea [62,70].
According to the results of a comparative analysis of dendrograms in a Korean province, cluster 11 accounted for the largest percentage, followed by clusters 10 and 7 [71]. According to a single-hospital study of children, the outbreaks were common between June and September in patients aged 8–16 years who presented with diarrhea (98%), fever (97%), abdominal pain (94 %), vomiting (37%), and headache (34%) [72].
The incidence of food poisoning by C. jejuni is gradually increasing [73], and 58 sequence types have been identified: ST-21 (24.2%), ST-50 (11.9%), ST4253, ST-51, and ST-4739 in domestic and overseas traveler diarrhea patients. Multilocus sequence typing (MLST) analysis identified 19 types of clone complexes (CCs), including CC21 (45.9%), CC443, CC22, and CC464, and sequence types within CC21 were ST21, ST-50, ST-4253, ST-19, ST-760, and ST-1811 [74].
5. Clostridium perfringens
Some C. perfringens strains produce C. perfringens enterotoxin (CPE), which is responsible for food poisoning, antibioticassociated diarrhea, and nosocomial diarrheal disease [75]. C. perfringens are classified into 5 types (A, B, C, D, E), depending on the produced toxins; those that produce CPE are mostly classified into type A, but they can also be types C and D [76]. CPE is a toxin that can cause food poisoning and intestinal diseases, and the toxin gene is identified as chromosomal enterotoxin (C-cpe) or plasmid-borne cpe gene (P-cpe) [77]. The disease phenotypes differ depending on the cpe gene locus [78].
Between 2014 and 2015, the location of the cpe gene and MLST type were analyzed in C. perfringens separated from sporadic acute diarrhea patients and mass food poisoning patients, and there was a difference in the genetic system between the 2 groups. However, ST-41 is a P-cpe that is commonly separated from the 2 groups and believed to be able to cause diarrhea and food poisoning [79].
6. Bacillus cereus
According to KCDC data from 2005–2007, the monthly detection rate distribution of B. cereus was similar to that of pathogenic intestinal bacteria, such as pathogenic E. coli, Salmonella spp., and S. aureus, which showed a similar pattern in subsequent studies. In 2008, through the analysis of mass food poisoning patients and ingested grains, the emetic type gene was first identified among the diarrheal toxin and emetic toxin genes [80].
In an analysis of the stool samples of patients with acute diarrheal illness from 2012–2015, the isolation rate of B. cereus was around 10% in children under 10 years of age, while the distribution according to the toxin gene was 80% and 20% for diarrheal and emetic toxin genes, respectively. The detection rate of toxin genes, such as enFM, nheA, cytK2, hblC, and bceT, was lower than that of the diarrheal toxin strains in the vomiting toxin strains; enFM and nheA were the main toxin genes in the 2 strains [81].
7. Other bacteria
Prior to 2008, Shigella flexneri was common among Shigella spp. at about 30%, but Shigella sonnei has become common since then, and the detection rate of Shigella spp. has gradually decreased [82]. Shigella spp. occurred sporadically but was mainly separated in November and December, showing a decrease, with a low separation rate among groups with patients under 10 years of age and a detection rate of less than 1% in a recent analysis [83].
The strain types analyzed by MLST were 11 sequence types, including ST-3, ST-332, and ST-8, and were not related to each other, indicating that the domestically separated Vibrio parahaemolyticus was genetically diverse. The most common sequence type was ST-3, and ST-1530, and ST-1531 were identified only in Korea. The largest CCs were CC-3 and CC-332, and most of the toxin gene analyses showed that they had tdh or trh genes [84].
Yersinia pseudotuberculosis was not included in the KCDC monitor, but a number of pediatric patients who were hospitalized with symptoms such as fever, digestive symptoms, and renal failure at a university hospital tested positive for Yersinia immunoglobulin A immediately after symptom onset, and Yersinia infection was confirmed in May 2017 [85].
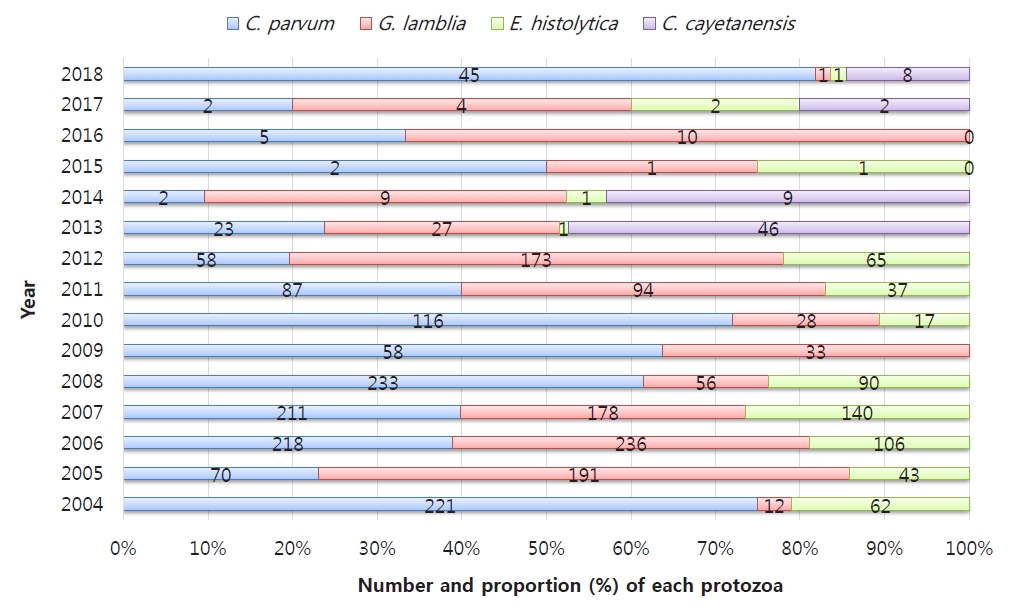
Protozoal infection (Fig. 3)
The 4 types of protozoa, which cause nationally notifiable infectious diseases, Cryptosporidium parvum, Giardia lamblia, Entamoeba histolytica, and Cyclospora cayetanensis, act as causative pathogens for acute diarrheal illness, especially a waterborne infection, which can be life-threatening in cases of infection in immunodeficient patients or young infants [86-88].
From 2004 to 2006, the KCDC reports on gastroenteritis caused by protozoa and bacterial and viral duplication showed a detection rate of about 1.4% in children under 5 years of age, followed by G. lamblia, C. parvum, and E. histolytica [86]. C. parvum was the most common of all protozoa and more common in children than in adults, while the frequency of G. lamblia gradually decreased in a later study [86,89].
Since the introduction of a new test method for protozoa in 2012, the sensitivity has increased from 4.32% in the previous year to 11.24%; during this time, G. lamblia, C. parvum, and E. histolytica were detected in order, but C. cayetanensis was not identified [90]. C. cayetanensis was the most common in 2013 [88], but G. lamblia was the most common in the overall statistics from 2009–2014 [80]. The 2015–2018 survey showed that the overall detection rate of protozoa decreased to less than 1%, but that of C. parvum, which began to increase after 2018, was the highest at about 64%, apparently due to improving efficiency of the preconditioning process for diagnosis [87].
Between 2009 and 2014, the distribution by age was followed by those aged 60 years or older with a detection rate of about 3.6 % for those under 10 years of age, and the seasonal distribution was higher in May and October between seasons [60]. According to a survey conducted in 2015–2018, about 50% of all patients were under 10 years of age [87].
Similar outbreaks of mass infection as in other developed countries have been reported in Korea, including G. lamblia in 2010 and C. parvum through the pollution of drinking water sources due to an aging water supply and sewage contamination in 2012 [88]. The detection of Cryptosporidium and Cyclospora from raw vegetables, such as perilla leaves, winter-grown cabbages, chives, sprouts, blueberries, and cherry tomatoes, suggests that monitoring and prevention systems should be used to prevent protozoal infections [91].
A subtype survey conducted for the first time in the KCDC investigation in 2013 identified C. parvum subtypes IIe and IIa and G. lamblia assemblage A type, and the similarity in genetic subtypes between humans and animals suggest zoonosis. The results of a molecular epidemiological analysis conducted between 2013 and 2016 in patients with acute diarrhea revealed that C. parvum belonged to the IIa family and was subtyped as IIaA13G2R1, IIaA14G2R1, IIaA15G2R1, and IIaA18G3R1, while the Giardia duodenalis genotype was identified as assemblage A.92) The presence of C. hominis infection was identified by real-time polymerase chain reaction–restriction fragment length polymorphism [93].
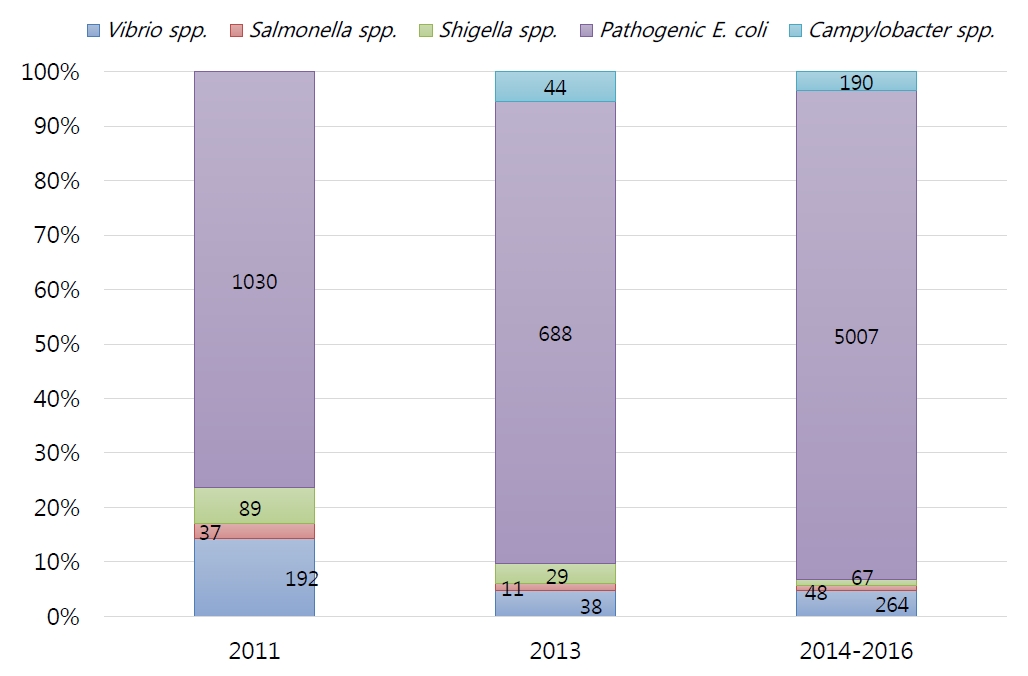
Foreign traveler’s diarrhea (Fig. 4)
The results of the 2011 survey by the Incheon International Airport Quarantine Service confirmed that various types of diarrhea-causing bacteria have been introduced from 14 countries; of them, about 80% were bacterial infections, although they vary among regions.
In 2011, the detection rate of bacterial pathogens in patients with acute AGE was 19%, and pathogenic E. coli (76%) was the most common bacterium, followed by Vibrio spp. (14%), Shigella spp. (7%), and Salmonella spp. (3%), and ETEC was the most common pathogenic E. coli spp [94]. Later, in a survey conducted in 2013, the detection rate of pathogenic bacteria from 16 countries surged to 61.2%, with the distribution of causative bacteria being similar [95].
In the 2014–2016 survey, the detection rate of pathogenic bacteria was 79.3%, followed by pathogenic E. coli (5,009 cases, 90%), Vibrio spp. (264 cases, 5%), Campylobacter spp. (190 cases, 3%), Shigellosis spp. (67 cases, 1%), and Salmonella spp. (48 cases, 1%). Of the pathogenic E. coli, EPEC was the most common (39%), followed by ETEC (36%), EAEC (25%), and EIEC (1%). Five cases (0.1%) of EHEC, a category I nationally notifiable infectious disease, were separately found over the 3 years.
According to the distribution of 37 countries from where patients with acute AGE arrived in Korea, 1,555 patients (22%) arrived from Vietnam, followed by 1,291 (18%) from China, 991 (14%) from Indonesia, 983 (14%) from Cambodia, 486 (7%) from Laos, and 362 (5%) from the United Arab Emirates. Outside Asian countries, Ethiopia had the largest number, with 8 patients (1.3%), followed by 27 (0.4%) from the United States, 23 (0.3%) from Kenya, and 18 (0.3%) from Brazil [96]. Among the Salmonella strains introduced from abroad, S. Anatum had the highest separation rate (12.1%). In addition to S. India, S. Bovismorbificans, and S. Poona, S. Wandsworth (an antigen is expressed in Salmonella I 39:b:1,2) and Salmonella I 4, [5],12:i:- were identified during the quarantine process in 2016.
Among the Salmonella strains introduced from abroad, S. Anatum (12%) was the most commonly identified, and in addition to S. India, S. Bovismorbificans, and S. Poona, S. Wandsworth, Salmonella I 4,[5],12:i:-, and S. Hvittingfoss were identified as rare serum types [97-99]. Plesiomonas shigelloides, a rare causative bacterium, was identified in 2015 [100].
Conclusions
The distribution of viruses and bacteria, the main causes of acute AGE, changes annually. Moreover, pathogens of different genotypes or those that were previously not reported or well known in Korea have been introduced due to an increase in overseas travel, international exchange, and foreigner influx. Therefore, it is necessary to continue researching, monitoring, and managing each pathogen. In addition, attention should be paid to increasing familiarity with the national reporting system.





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link PubMed
PubMed Download Citation
Download Citation