Article Contents
| Korean J Pediatr > Volume 56(6); 2013 |
Abstract
Purpose
In children with acute leukemia, bone marrow genetic abnormalities (GA) have prognostic significance, and may be the basis for minimal residual disease monitoring. Since April 2007, we have used a multiplex reverse transcriptase-polymerase chain reaction tool (HemaVision) to detect of GA.
Methods
In this study, we reviewed the results of HemaVision screening in 270 children with acute leukemia, newly diagnosed at The Catholic University of Korea from April 2007 to December 2011, and compared the results with those of fluorescence in situ hybridization (FISH), and G-band karyotyping.
Results
Among the 270 children (153 males, 117 females), 187 acute lymphoblastic leukemia and 74 acute myeloid leukemia patients were identified. Overall, GA was detected in 230 patients (85.2%). HemaVision, FISH, and G-band karyotyping identified GA in 125 (46.3%), 126 (46.7%), and 215 patients (79.6%), respectively. TEL-AML1 (20.9%, 39/187) and AML1-ETO (27%, 20/74) were the most common GA in ALL and AML, respectively. Overall sensitivity of HemaVision was 98.4%, with false-negative results in 2 instances: 1 each for TEL-AML1 and MLL-AF4. An aggregate of diseasesspecific FISH showed 100% sensitivity in detection of GA covered by HemaVision for actual probes utilized. G-band karyotype revealed GA other than those covered by HemaVison screening in 133 patients (49.3%). Except for hyperdiplody and hypodiploidy, recurrent GA as defined by the World Health Organizationthat were not screened by HemaVision, were absent in the karyotype.
In the recent diagnosis and classification of acute leukemia, genetic characterization, rather than morphology-based description, has played a critical role. A clear example of the emphasis on genetic abnormalities (GA) in the diagnosis of hematologic malignancies is the inclusion of acute leukemias with "recurrent genetic abnormalities," as a subdivision within the recent World Health Organization (WHO) classification of hematopoietic tumors1). Many of these GA have prognostic utility2,3), and may form the basis for minimal residual disease-based treatment4).
Methods for evaluation of GA include polymerase chain reaction (PCR)-based molecular methods, and cytogenetic studies including fluorescence in situ hybridization (FISH) tests and conventional G-band karyotyping. HemaVision (DNA Technology, Aarhus, Denmark) is a method of multiplex reverse transcriptase (RT)-PCR that allows for the rapid detection of 28 different GA that have been discovered in both acute and chronic leukemia. Since the initial report on the utility of this method of multiplex RT-PCR screening in acute leukemia5), several studies have confirmed the efficacy of this tool for rapid and sensitive evaluation of GA in a large number of patients6-8). Affirmative results of the application of the HemaVision tool in the Korean patient population have also been reported, with most of the patients tested consisting of adults9-11). However, since an early study on the applicability of screening using HemaVision in a cohort of children with acute myeloid leukemia (AML)12), little evaluation of this tool has been done in a solely pediatric leukemia patient group, and none, so far, has been undertaken in a Korean pediatric population. Considering that the overall incidence of recurrent GA may differ between pediatric and adult patients, and considering also that the relative incidence of a specific GA may differ according to the ethnicity of the patient group studied13), an evaluation of HemaVision in Korean children may yield unique results concerning the utility of this tool in detecting GA overall, as well as specific GA that may be of greater prognostic significance for children.
In this study, we aimed to determine the utility of HemaVision in the detection of GA in a cohort of children diagnosed with acute leukemia at the Department of Pediatrics, Seoul St. Mary's Hospital, The Catholic University of Korea College of Medicine.
We have utilized HemaVision for the screening of GA in patients with acute leukemia since April, 2007. Our study cohort consisted of children newly diagnosed with acute leukemia at the Department of Pediatrics, The Catholic University of Korea from April 2007 to December 2011. Patients who had initially been diagnosed at another institution and had received previous chemotherapy before referral to our institution were excluded from the study cohort. Pertinent patient information, such as age at diagnosis, gender, and immunophenotype were retrospectively reviewed from medical records. All patients underwent HemaVision screening, and cytogenetic tests including an aggregate of disease-appropriate FISH tests and G-band karyotyping with bone marrow (BM) samples at the time of diagnosis. The study design received approval from the Institutional Review Board (KC12RISI0679).
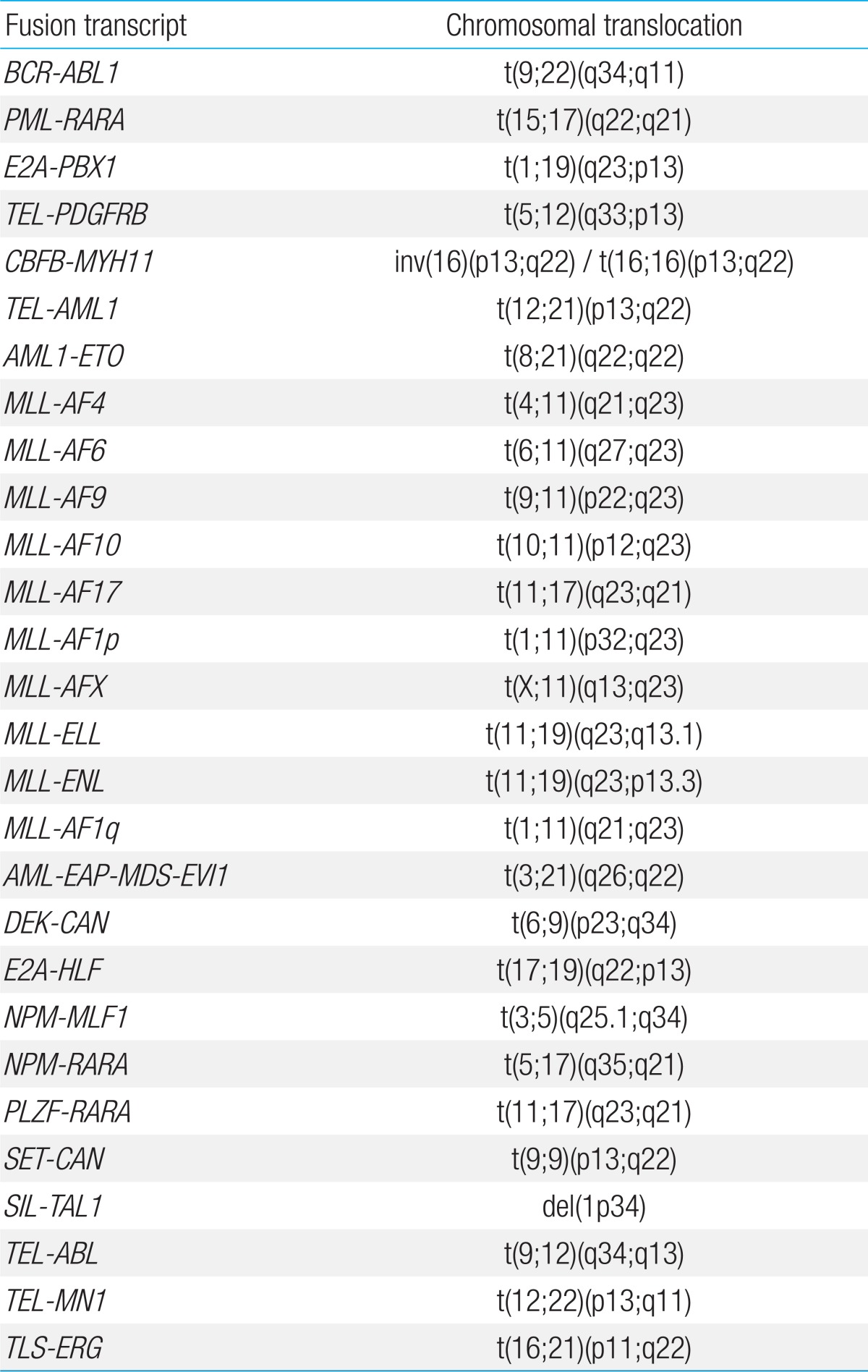
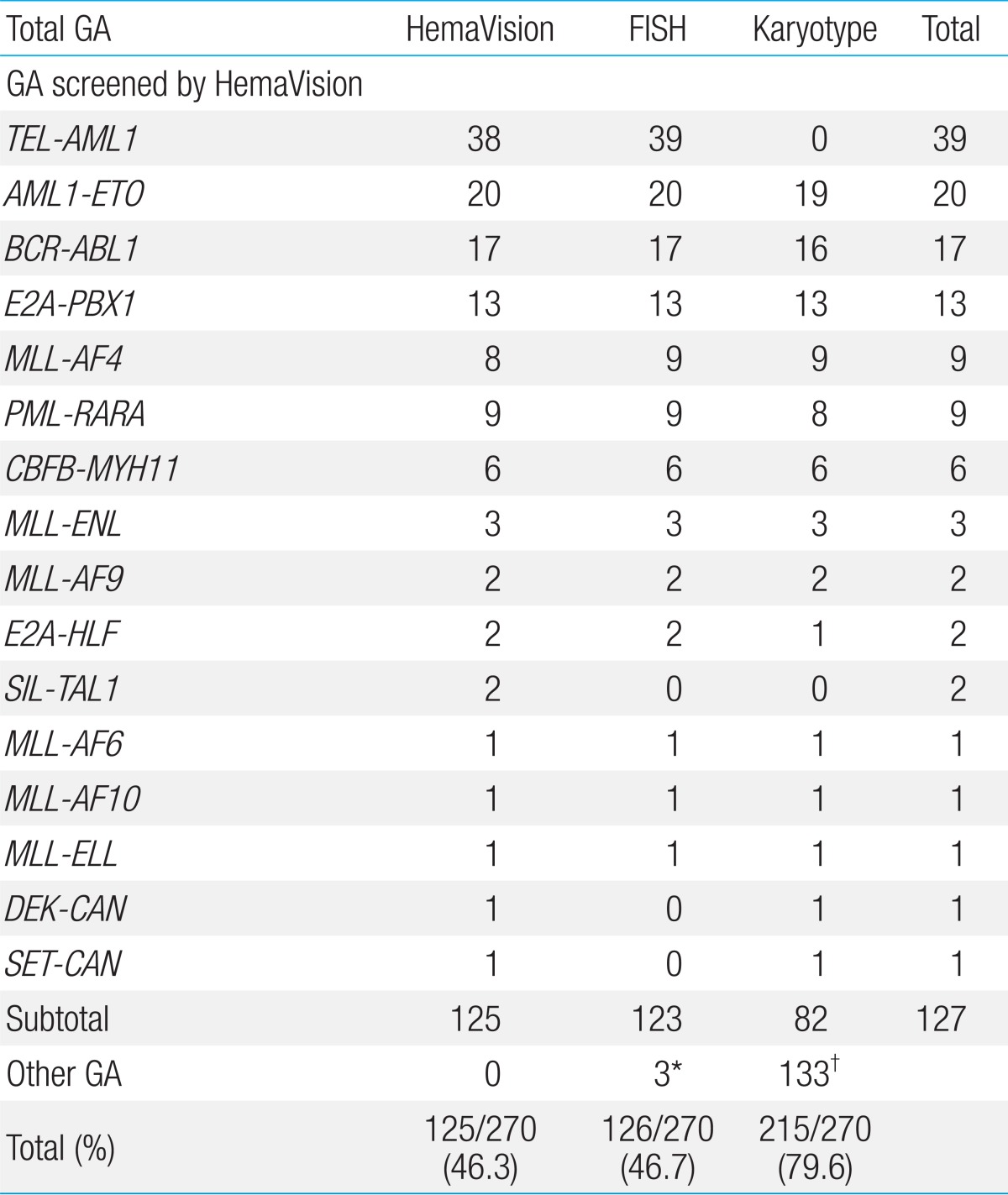
RNA was extracted from BM samples using High Pure RNA Isolation Kit (Roche Diagnostics, Mannheim, Germany). Multiplex RT-PCR with HemaVision was done according to manufacturer's instructions, with performance of 2 sequential nested PCR reactions, details of which have been published in a previous report on HemaVision application6). The 28 GA screened for by HemaVision are shown in Table 1.
An aggregate of interphase FISH tests were done according to patient disease. For acute lymphoblastic leukemia (ALL), the following studies were done: BCR-ABL1, TEL-AML1, C-MYC, MLL, TCF3. For AML, the following studies were done: BCR-ABL1, AML1-ETO, PML-RARA, MLL, CBFB, 5q, 7q31. The probes used were as follows: LSI BCR/ABL1, RUNX1/RUNX1T1, PML/RARA dual color, dual fusion translocation probe, LSI ETV6/RUNX1 extra signal dual color, translocation probe, LSI MYC, MLL, CBFB dual color, break apart rearrangement probe, LSI EGR1 (5q31)/D5S23 probe, D7S486 (7q31)/CEP7 probe (Abbott Laboratories. Abbott Park, IL, USA), and E2A break apart LPS 019 probe (Cytocell Ltd., Cambridge, UK). The BM cells from routine chromosomal preparations were used. Pretreatment and hybridization were performed, in accordance with the manufacturer's recommendation. Results were considered abnormal if the percentage of nuclei with abnormal signals exceeded the normal reference ranges. For the purposes of this study, except for 5q, 7q31 probes, a FISH test was considered positive if the test aided in the identification of a specific translocation, rather than incidental numerical abnormalities. The 5q and 7q31 tests were considered positive if monosomies or arm deletions of chromosomes 5 or 7 were identified.
Chromosomal analysis was performed at diagnosis on short-term cultures with BM cells at metaphase, according to a conventional G-banding analysis using standard cytogenetic protocols. At least 20 metaphases were analyzed in each case, and the clonal abnormalities were classified according to International System for Chromosome Nomenclature 2009 guidelines14).
The overall incidence of GA, detected either through HemaVision, FISH or G-band karyotyping, in the patient cohort was evaluated, as well as the incidence of GA within each leukemia type and the positivity rates of each detection method.
The GA screened by HemaVision include many recurrent abnormalities that have prognostic significance. The incidence of these recurrent GA within the overall cohort, regardless of diagnostic method, and within each leukemia type was determined.
The positivity rate of HemaVision within each leukemia type, and the overall sensitivities of HemaVision and FISH testing in detecting GA were assessed.
As G-band karyotype analysis may reveal additional information concerning numerical and structural chromosomal abnormalities for patients with positive HemaVision tests, the presence of such abnormalities in the BM karyotype was noted. Karyotyping may also show GA other than those covered by HemaVision screening, and the number of patients with such GA was also determined.
The GA screened by HemaVision coincide significantly with the recurrent GA of both ALL and AML that are part of the 2008 WHO classification1). However, there are several categories in the WHO classification that are not screened by HemaVision: hyperdiploidy, hypodiploidy, t(5;14)(q31;q32) for ALL, and inv(3)(q21q26.2) or t(3;3)(q21;q26.2), t(1;22)(p13;q13) for AML. Our final study objective was to evaluate for the incidence of these GA not screened by HemaVision in the karyotype analysis to observe whether initial screening with HemaVision allows for appropriate patient categorization according to WHO classification for recurrent GA.
Overall, 270 patients (153 males, 117 females) were newly diagnosed with acute leukemia at our institution during the study period. This cohort consisted of 187 ALL (69.3%), 74 AML (27.4%), 8 mixed phenotype (MPAL) (3%), and 1 acute undifferentiated leukemia patient (0.4%). Amongst the 187 patients with ALL, 167 were found to have precursor B cell ALL according to immunophenotype, while the remaining 20 patients were diagnosed with T cell ALL. Median age at diagnosis for the study cohort was 6.8 years (range, 0.2 to 18.8 years).
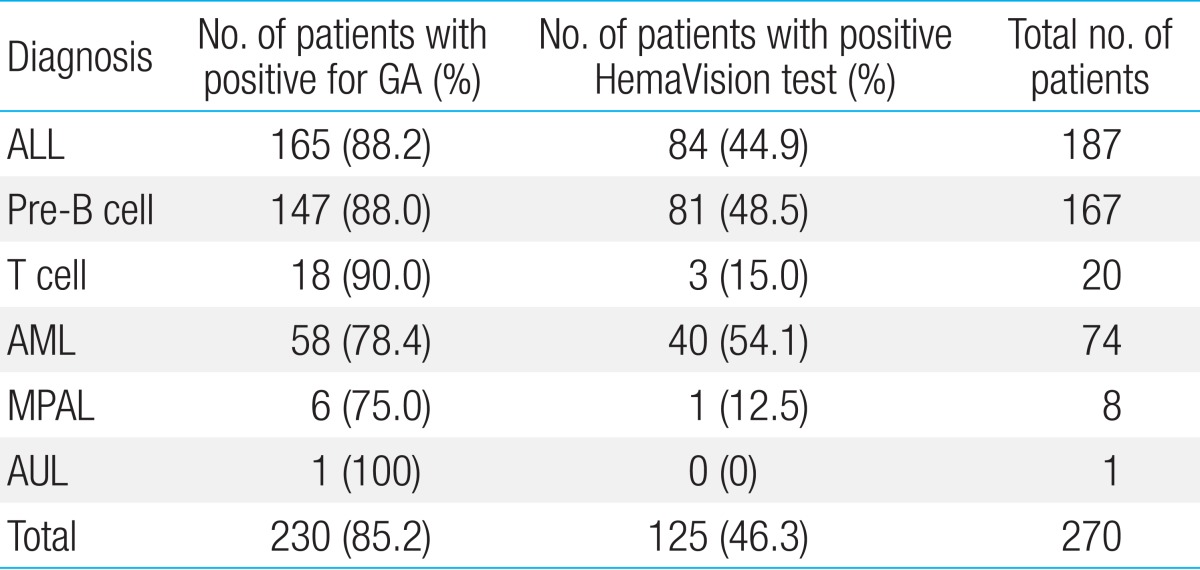
Within the entire cohort, 230 patients (85.2%, 230/270) were found to have GA either through HemaVision, FISH or karyotype study. Forty patients had normal karyotype, and had negative findings for both HemaVision and FISH.
With regards to each leukemia type, GA was found in 88.2% of ALL patients (165/187), 78.4% of AML patients (58/74), and 75.0% of MPAL patients (6/8) (Table 2). Of patients with precursor B ALL, 88.0% of patients (147/167) were diagnosed with GA, while 18 of 20 patients (90%) were found to have GA in the T cell leukemia subgroup.
HemaVision screening was positive for 16 of 28 possible GA in 46.3% of patients (125/270), and FISH in 46.7% (126/270) of patients (Table 3). G-band karyotype revealed GA in 215 patients (79.6%, 215/270) and proved to have the highest positivity rate in terms of detecting GA of all 3 tests.
Overall, 127 patients were diagnosed with recurrent GA that were incorporated into HemaVision screening, through any of the 3 test methods. Within the ALL subgroup, 39 patients were found to have TEL-AML1, confirming this GA to be the most abnormality (20.9%, 39/187). Other recurrent GA diagnosed in the ALL subgroup were as follows: BCR-ABL1 (n=17, minor sub-type 15, major subtype 2), E2A-PBX1 (n=13), MLL-AF4 (n=9), MLL-ENL (n=2), E2A-HLF (n=2), and MLL-AF9 (n=1) for patients with precursor B ALL. Minor BCR-ABL1 subtypes included 14 patients with e1a2, and 1 patient with e1a3 GA. The 2 patients with major BCR-ABL1 subtype had b2a2 GA. For patients with T cell ALL, 3 patients were found to have recurrent GA, one patient each for the following: MLL-ENL, SET-CAN, and SIL-TAL1.
AML1-ETO (n=20) was the most common recurrent GA in the AML subgroup, with the remainder consisting of the following: PML-RARA (n=9), CBFB-MYH11 (n=6), MLL-AF6 (n=1), MLL-AF9 (n=1), MLL-AF10 (n=1), MLL-ELL (n=1), and DEK-CAN (n=1).
Amongst the 8 patients with MPAL, 1 patient with T/Myeloid phenotype was diagnosed with a SIL-TAL1 abnormality.
According to leukemia subtype, HemaVision screening was positive in 44.9% of ALL (84/187), 54.1% of AML (40/74), and 1 MPAL patient (Table 2) for a total of 125 positive patients. HemaVision was positive in 50.9% of ALL patients with any GA (84/165), and 70% of AML patients with any GA (40/58).
The overall sensitivity of HemaVision screening was 98.4% (125/127). Of the 2 patients with GA not detected by HemaVision were 1 patient with MLL-AF4 confirmed by both FISH and karyotype, and 1 patient with TEL-AML1 confirmed by FISH alone. A subsequent review of these 2 patients revealed that splicing site differences between the fusion transcript detected by HemaVision and the actual transcript found in the patients were the main reason for the failure of HemaVision screening.
FISH testing showed 100% sensitivity (123/123) in the detection of GA covered by HemaVision for actual probes utilized. Besides these recurrent GA, testing with FISH (7q31) allowed for the diagnosis of 2 AML patients with monosomy 7 and 1 AML patient with del(7q) (Table 3).
Of the 125 HemaVision positive patients, the karyotype revealed either additional numerical or structural abnormalities in 78 patients (62.4%), including 20 (16%) with additional numerical abnormalities only, 35 (28%) with additional structural abnormalities only, and 23 (18.4%) with both additional numerical and structural abnormalities. Of the 270 patients in the overall cohort, 133 (49.3%) had numerical or structural GA other than those covered by HemaVision.
Hyperdiploidy and hypodiploidy, both categorized as recurrent GA in ALL by WHO classification, were identified by G-band karyotype in 19.8% (37/187) and 1.1% (2/187) of ALL patients respectively. However, other recurrent GA defined by WHO that were not covered by HemaVision, (5;14)(q31;q32), inv(3)(q21q 26.2) or t(3;3)(q21;q26.2), and t(1;22)(p13;q13), were not found in karyotype analysis of the overall cohort.
Although the data of GA in childhood acute leukemia in this paper derived from a single Korean institution, several salient points can be made with regards to the incidence of specific GA. First, nearly 90% of ALL patients were found to have GA. TEL-AML1, overall the most common GA, was detected in 20.9% of ALL patients, indicating an incidence that was similar to figures previously reported from Western countries15). Incidence of GA was also high in our AML patients (78.4%). A noteworthy point with regards to the incidence of specific GA in our AML patients is the significance of AML1-ETO as the most common GA (27%, 20/74), in contrast to past studies where incidence of this abnormality ranged from 9-12%16,17). Another clear difference is the low rate of combined MLL gene rearrangements in our AML cohort (5.4%, 4/74), whereas previous studies have noted an approximately 20% incidence for this GA. Prognostic implications of these disparities are evident, as detection of AML1-ETO predicts a better response to treatment. Although the small number of AML patients included in this study do not allow for definite conclusions, discrepancies in specific GA incidence hint at the possibility of population-based differences that may become clear with evaluation of a greater number of Korean AML children.
Overall, 46.3% of patients in the cohort had GA identified through HemaVision, a figure which is higher than the 20-30% positivity rates reported from cohorts consisting mostly of adult patients8,11). Other studies have analyzed the positivity rate of HemaVision within their respective pediatric and adult subcohorts and found the rate to be higher in children6,10). The relatively high incidence of GA detected by HemaVision in our study may be further evidence of the greater diagnostic significance of this tool in children with leukemia, especially AML patients in whom HemaVision was positive in 70% of patients with any GA, in contrast to the 50.9% positivity rate amongst ALL patients with any GA. A note should be made, however, concerning the unequal contributions made by each component of HemaVision to the overall high detection rate. The 3 most common GA detected by HemaVision, TEL-AML1, AML1-ETO, BCR-ABL1, comprised more than half of all positive cases. Twelve of 28 GA were not represented at all in our cohort. Hence, the high positivity rate for HemaVision may stem mostly from high incidence rates of select GA within our cohort.
Besides the relatively high positivity rate and sensitivity shown in our study, previously known advantages of HemaVision include the ability to detect cryptic translocations. In the pediatric setting, one of the most important cryptic translocations in terms of prognosis would be TEL-AML1 which is not detected in karyo-type study. Also, in contrast to karyotyping which is a lengthy process, HemaVision allows for the rapid identification of important GA. Finally, the RT-PCR method may be deemed cost-effective, as it allows for the screening of 28 GA at 2-3 times the cost of one FISH test, according to current domestic pricing.
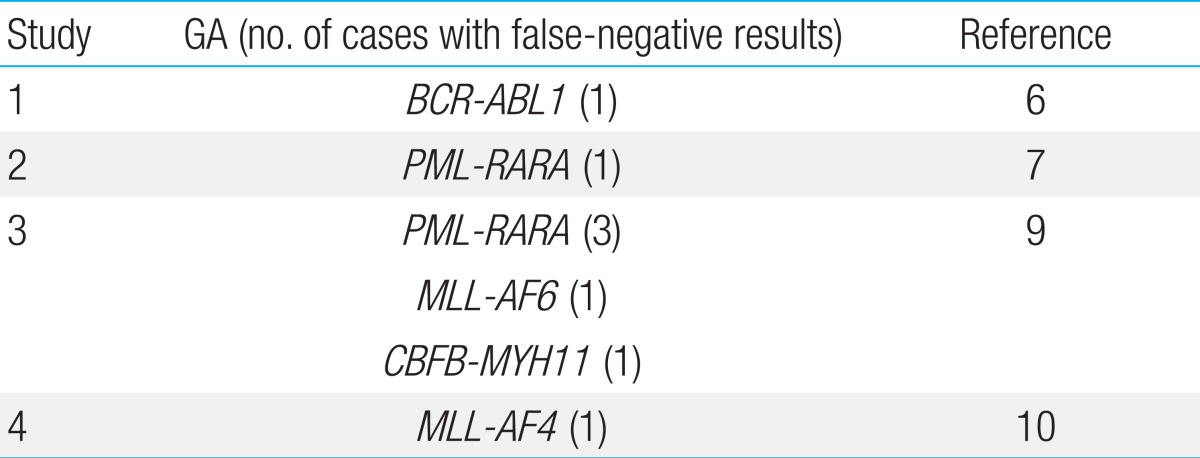
HemaVision screening proved to be falsely negative on 2 occasions: one patient each with TEL-AML1 and MLL-AF4, both GA with well-known prognostic significance. In Table 4, we have summarized the false-negative results of HemaVision that have been reported in the literature. Reasons for false-negative results may include differences in splicing sites in the creation of the fusion transcript, and lack of mRNA for adequate testing. For MLL gene rearrangements, despite the variety of fusion transcripts including MLL that are screened for by HemaVision, the diversity of MLL partners even within one particular chromosome (for example, the numerous fusion partners for MLL on 17q11,12)) may elude identification with HemaVision. In our study, both false-negative GA were identified through FISH. Hence, confirmation of HemaVision results with empirical use of an aggregate of FISH tests, including MLL and those that test GA for which literature has reported false-negative HemaVision results, may lead to the most accurate genetic diagnosis of a leukemia patient.
G-band karyotyping allowed for diagnosis of either additional numerical or structural abnormalities in 62.4% of patients with positive HemaVision tests. The prognostic implications of additional abnormalities in patients with recurrent GA, as defined by WHO, remain unresolved, with several studies concluding that additional cytogenetic abnormalities do not alter the prognosis conferred by a recurrent GA18,19). Nevertheless, the identification of additional abnormalities in the karyotype forms the basis for further studies on their prognostic relevance in the context of concurrent GA. Furthermore, karyotype study allowed for the identification of GA other than those screened by HemaVision in nearly half the patients in the cohort, including monosomies and arm deletions which may negatively affect the outcome of AML patients. Hence, the G-band karyotype remains an essential component for the comprehensive genetic evaluation of the acute leukemia patient.
Screening using HemaVision did not allow for full categorization according to the 2008 WHO classification for recurrent GA of ALL and AML, as HemaVision did not include 5 of these abnormalities. However, besides hyperdiploidy and hypodiploidy of ALL which were identified in karyotype study, none of the remaining 3 abnormalities, that was, t(5;14)(q31;q32), inv(3)(q21q26.2), and t(1;22)(p13;q13), were found in our cohort. Hence, in our single institution cohort, screening with HemaVision allowed for full genetic assessment according to WHO criteria, except for numerical abnormalities which could not be tested using an RT-PCR based tool. In addition, our study raises the question of whether the WHO classification of recurrent abnormalities fully reflects the incidence of important genetic aberrations in a Korean pediatric population. Of the 3 abnormalities that were absent in our cohort, t(1;22)(p13;q13) is known to occur almost solely in infants and young children20), and a nationwide study investigating its incidence in Korean AML children may shed greater information on its domestic importance.
In conclusion, our experience of screening de novo pediatric acute leukemia patients for GA with HemaVision proved to be both useful, as 46% of the overall cohort had a positive test, and sensitive. Higher positivity rate in our cohort compared to previous results derived from mostly adult patient-based cohorts emphasize the utility of this tool in childhood leukemia. Greater diagnostic accuracy, with relevant prognostic consequences for each individual patient, may be achieved by HemaVision screening complemented by an aggregate of disease-specific FISH tests, especially including MLL and other GA for which false-negative HemaVision results have been reported.
Acknowledgments
We are grateful to the staff and personnel of the Department of Laboratory Medicine, Seoul St. Mary's Hospital, The Catholic University of Korea, for accurate diagnosis of our patients.
References
1. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, , editors. WHO classification of tumors of haematopoietic and lymphoid tissue. 4th ed. Lyon: IARC, 2008.
3. Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, et al. The Medical Research Council Adult and Children's Leukaemia Working Parties. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. Blood 1998;92:2322ŌĆō2333.


4. Pallisgaard N, Clausen N, Schroder H, Hokland P. Rapid and sensitive minimal residual disease detection in acute leukemia by quantitative real-time RT-PCR exemplified by t(12;21) TEL-AML1 fusion transcript. Genes Chromosomes Cancer 1999;26:355ŌĆō365.


5. Pallisgaard N, Hokland P, Riishoj DC, Pedersen B, Jorgensen P. Multiplex reverse transcription-polymerase chain reaction for simultaneous screening of 29 translocations and chromosomal aberrations in acute leukemia. Blood 1998;92:574ŌĆō588.


6. Olesen LH, Clausen N, Dimitrijevic A, Kerndrup G, Kjeldsen E, Hokland P. Prospective application of a multiplex reverse transcription-polymerase chain reaction assay for the detection of balanced translocations in leukaemia: a single-laboratory study of 390 paediatric and adult patients. Br J Haematol 2004;127:59ŌĆō66.


7. Hutchings Hoffmann M, Wirenfeldt Klausen T, Hasle H, Schmiegelow K, Brondum-Nielsen K, Johnsen HE. Multiplex reverse transcription polymerase chain reaction screening in acute myeloid leukemia detects cytogenetically unrevealed abnormalities of prognostic significance. Haematologica 2005;90:984ŌĆō986.

8. Meyer-Monard S, Parlier V, Passweg J, Muhlematter D, Hess U, Bargetzi M, et al. Combination of broad molecular screening and cytogenetic analysis for genetic risk assignment and diagnosis in patients with acute leukemia. Leukemia 2006;20:247ŌĆō253.


9. Park JS, Yi JW, Jeong SH, Lee HW, Kang SY, Choi JH, et al. Comparison of multiplex reverse transcription polymerase chain reaction and conventional cytogenetics as a diagnostic strategy for acute leukemia. Int J Lab Hematol 2008;30:513ŌĆō518.


10. Choi HJ, Kim HR, Shin MG, Kook H, Kim HJ, Shin JH, et al. Spectra of chromosomal aberrations in 325 leukemia patients and implications for the development of new molecular detection systems. J Korean Med Sci 2011;26:886ŌĆō892.



11. Song MJ, Kim HJ, Park CH, Kim SK, Ki CS, Kim JW, et al. Diagnostic utility of a multiplex RT-PCR assay in detecting fusion transcripts from recurrent genetic abnormalities of acute leukemia by WHO 2008 classification. Diagn Mol Pathol 2012;21:40ŌĆō44.


12. Strehl S, Konig M, Mann G, Haas OA. Multiplex reverse transcriptase-polymerase chain reaction screening in childhood acute myeloblastic leukemia. Blood 2001;97:805ŌĆō808.


13. Sazawal S, Bhatia K, Gutierrez MI, Saxena R, Arya LS, Bhargava M. Paucity of TEL-AML 1 translocation, by multiplex RT-PCR, in B-lineage acute lymphoblastic leukemia (ALL) in Indian patients. Am J Hematol 2004;76:80ŌĆō82.


14. Shaffer LG, Tommerup N. ISCN 2005: an international system for human cytogenetic nomenclature (2005). Basel: Karger, 2005.
15. Borkhardt A, Cazzaniga G, Viehmann S, Valsecchi MG, Ludwig WD, Burci L, et al. Associazione Italiana Ematologia Oncologia Pediatrica and the Berlin-Frankfurt-M├╝nster Study Group. Incidence and clinical relevance of TEL/AML1 fusion genes in children with acute lymphoblastic leukemia enrolled in the German and Italian multicenter therapy trials. Blood 1997;90:571ŌĆō577.


16. Raimondi SC, Chang MN, Ravindranath Y, Behm FG, Gresik MV, Steuber CP, et al. Chromosomal abnormalities in 478 children with acute myeloid leukemia: clinical characteristics and treatment outcome in a cooperative pediatric oncology group study-POG 8821. Blood 1999;94:3707ŌĆō3716.

17. Forestier E, Heim S, Blennow E, Borgstrom G, Holmgren G, Heinonen K, et al. Cytogenetic abnormalities in childhood acute myeloid leukaemia: a Nordic series comprising all children enrolled in the NOPHO-93-AML trial between 1993 and 2001. Br J Haematol 2003;121:566ŌĆō577.


18. Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood 2002;100:4325ŌĆō4336.





 PDF Links
PDF Links PubReader
PubReader PubMed
PubMed Download Citation
Download Citation