MicroRNAs as novel biomarkers for the diagnosis and treatment of pediatric diseases
Article information
Abstract
MicroRNAs (miRNAs) are highly conserved noncoding RNAs that regulate gene expression by silencing or degrading messenger RNAs. Many of the approximately 2,500 miRNAs discovered in humans are known to regulate vital biological processes, including cell differentiation, proliferation, apoptosis, and embryonic tissue development. Aberrant miRNA expression may have pathological and malignant consequences. Therefore, miRNAs have emerged as novel diagnostic markers and potential therapeutic targets for various diseases. Children undergo various stages of growth, development, and maturation between birth and adulthood. It is important to study the role of miRNA expression in normal growth and disease development during these developmental stages. In this mini-review, we discuss the role of miRNAs as diagnostic and prognostic biomarkers in various pediatric diseases.
Key message
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression post transcriptionally, and MiRNA expression levels vary with developmental stages. MiRNAs play an important role in several biological processes in children, including growth, neuro-development, inflammation, and tumor formation. Research on miRNAs may uncover the molecular mechanisms underlying various pediatric diseases, leading to the development of novel biomarkers that aid in the diagnosis, treatment, and prognosis of these diseases.
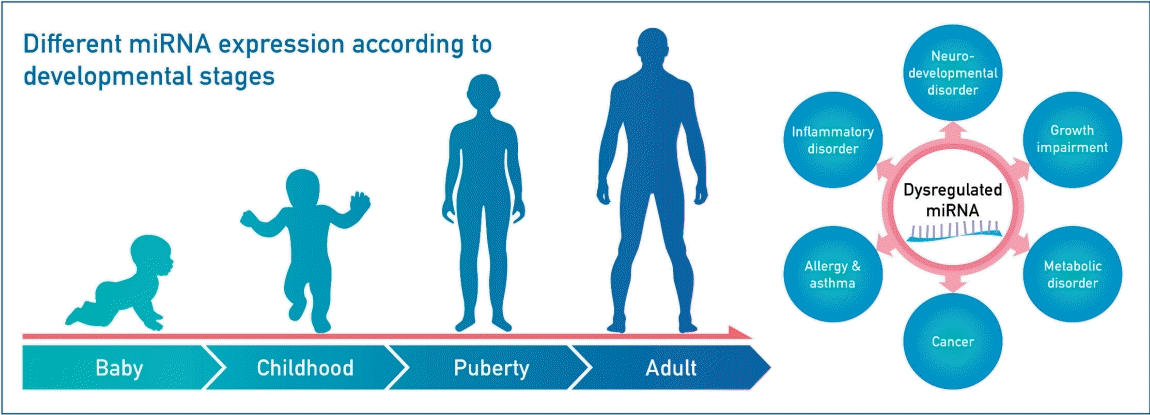
Graphical abstract. MicroRNAs (miRNAs) are involved in the expression of genes that regulate normal human growth and development. Therefore, their dysregulation may lead to several diseases. miRNA research is important because miRNAs could serve as novel biomarkers for the diagnosis, progression, and prognosis of pediatric diseases.
Introduction
MicroRNAs (miRNAs) are small, noncoding, single-stranded RNAs (18–23 nucleotides) that regulate posttranscriptional silencing of target genes through translational repression or degradation of the target messenger RNA (mRNA) [1]. MiRNAs were first discovered in 1993 in a study that demonstrated that lin-4, a regulator of larval developmental timing in the nematode Canenorbabditis elegans, does not code a protein, instead producing a pair of small RNAs [2]. It was subsequently found that the lin-4 RNAs had antisense complementarity with multiple sites in the 3’UTR of lin-14 gene, and could significantly reduce the quantity of LIN-14 protein [3]. These miRNAs have been shown to be conserved in several species, including humans. An increasing number of miRNAs have since been identified in animals, plants, and even viruses. These miRNAs regulate various biological processes, including developmental timing, cell death, cell proliferation, hematopoiesis, and nervous system patterning [4].
MiRNAs are abundant in several cell types, as well as in tissues and the circulation. Circulating miRNAs are released into body fluids, including blood, saliva, urine, breast milk, tears, and cerebrospinal fluid [5-7]. They can also be stably transported within extracellular vesicles, including apoptotic bodies, microvesicles, and exosomes. These vesicles are important for cell-to-cell communication because miRNAs can be selectively packaged, transported, and modulated for specific recipient cells [8,9]. MiRNA expression levels may differ among different tissues and organs, and between physiological and pathological states. Dysregulated miRNA expression is related to several pathologies, including cardiovascular diseases, epilepsy, metabolic syndrome, cancer, allergies, and autoimmune diseases. Changes in miRNA levels can explain phenotypic differences and provide information about the underlying genetic pathways. Furthermore, miRNA analysis is advantageous because the molecules are highly resistant to degradation, and their expression levels can be assessed within a few hours using small biological samples. Since miRNAs are stable and easily measurable, they can be used as diagnostic and prognostic biomarkers, as well as therapeutic targets [10,11].
An increasing number of studies have investigated the role of miRNAs in pediatric diseases, but there is still a need for further research in this area. Although several studies have reported the efficacy of miRNAs as biomarkers, most of these studies were performed in adults, and the findings may not be applicable to children because of the role of ontogeny in disease evolution and the therapeutic response [12]. A noninvasive and convenient testing method is required for younger children. Therefore, the present study reviews miRNA research and discusses their potential role as biomarkers in pediatric diseases.
MiRNA biogenesis
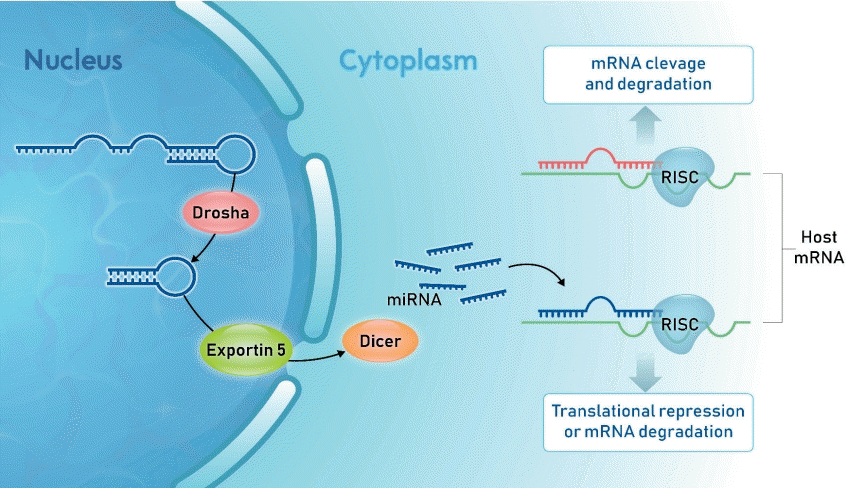
In the nucleus, RNA polymerase II (Pol II) transcribes miRNA genes into long hairpin structures called primary miRNA (Fig. 1). DGCR8 and Drosha are RNA endonucleases that cleave the nucleotide from the primary miRNA hairpin base to generate shorter precursor miRNA (premiRNA).
The nuclear pre-miRNAs are translocated by carrier protein Exportin-5 to the cytoplasm, where miRNA maturation occurs. Dicer ribonuclease with transactivation response element RNA-binding protein cuts away the loop of pre-miRNA to generate a mature miRNA duplex. This duplex consists of a guide strand (miRNA) and passenger strand (miRNA*) separated by a helicase. After miRNA* removal, mature miRNA is selected by the Argonaute 2 protein and incorporated into the RNA-induced silencing complex. Finally, the silencing complex binds to the mRNA molecule, usually at the 3’UTR of target mRNA, via sequence complementarity, resulting in cleavage degradation or translational repression of the mRNA [1,13,14].
A single miRNA may potentially pair with hundreds of different mRNAs. Single miRNAs can pleiotropically target hundreds or thousands of genes and perform organor cell-specific functions [11]. Similarly, each mRNA may be regulated by multiple miRNAs [15]. This multitargeting property of miRNAs is advantageous for disease modification because of the possibility of disrupting several pathological processes simultaneously. However, this also increases the potential for unanticipated side-effects in miRNA-based therapies [16]. It is estimated that miRNAs regulate the expression of approximately 60% of human genes [17].
Age-related miRNA expression
MiRNA expression varies according to the developmental stage. Age-related miRNAs may be involved in biological pathways related to growth and development in children. Huen et al. [18] analyzed miRNA expression in Mexican-American children between birth and the age of 7 years. They found that, in contrast with the decrease in miRNA expression seen in the peripheral blood with age in adults [19], the expression increased from birth to mid-childhood. Burgess et al. [20] analyzed miRNA expressions in fetal, pediatric, and adult livers, and found that hepatic miRNA expression changes with age, particularly between the fetal and pediatric stages. They suggested that miRNA expression may contribute to the clinical variability seen in hepatic drug metabolism. Lai et al. [21] compared peripheral blood miRNA expression levels between preterm infancy and adulthood. About one-third of the miRNAs were constantly expressed from the postnatal period to adulthood, one-third were differentially expressed between preterm infants and adults, and the remaining one-third were not detectable in either sample. Although it may be difficult to generalize the results to other racial groups, they nevertheless improve our understanding of the relationship of peripheral blood miRNA expression changes with postnatal development and aging.
MiRNAs and growth
MiRNAs are involved in bone formation, including skeletal development, longitudinal bone growth, and chondrocyte proliferation and differentiation [22]. MiRNAs are also critical for the regulation of hypothalamus function and pituitary development. They are known to be involved in growth hormone (GH) secretion and signaling pathways, including GH receptor (GHR) and insulin-like growth factor (IGF). Most of the evidence is based on animal models and in vitro studies, but there has been a recent increase in human studies, often overlapping with animal studies. Elzein and Goodyer [23] reported that human GHR expression is regulated by miRNA-129–5p, miRNA-142–3p, miRNA-202, and miRNA-16, which reduce GHR mRNA and protein in human cancer cell lines and control cells. Among these miRNAs, miRNA-202, and miRNA-16 are thought to regulate several aspects of the GH/IGF-1 axis. MiRNA-9 and miRNA-486 directly target IGF-1 3’UTR, leading to decreased IGF-1 expression at the mRNA and protein levels [24,25]. Gaoet al. [26] reported that most oral squamous cell carcinoma patients had significantly reduced let-7b expressions; let-7b suppresses cell growth by targeting IGF-1R and insulin receptor substrate-2.
An increasing number of studies on the association of the miRNA profile with growth impairment are being performed. A study of age-related serum miRNAs in untreated adult congenital GH-deficient patients demonstrated their involvement in insulin production, inflammation, and aging [27]. An investigation of plasma miRNA expression in Chinese idiopathic short stature children demonstrated significant upregulation of miRNA-185 expression and downregulation of miRNA-497 expression [28]. Mas-Parés et al. [29] studied umbilical cord miRNA profiles associated with catch-up growth in small for gestational age (SGA) children. Umbilical cord miRNA-576-5p was associated with catch-up growth and cardio-metabolic risk in SGA children, indicating that it could serve as a novel biomarker for catch-up growth in SGA infants. A recent study by our group of the exosomal miRNA profile of SGA children also highlighted their potential role as prognostic biomarkers for catch-up growth in SGA children [30].
Role of MiRNAs in diabetes and obesity
Several studies have reported miRNA dysregulation in patients with diabetes, obesity, and metabolic syndrome. The potential of miRNAs as biomarkers for diabetes and metabolic syndrome is being increasingly recognized [31-33]. Although studies comparing type 1 diabetes (T1D) patients with healthy controls have reported different miRNA expression signatures, but miRNA-21, miRNA-24, miRNA-148a, miRNA-181a-5p, miRNA-210–5p upregulation in T1D has been demonstrated in multiple independent studies [34]. MiRNAs are associated with pancreatic β-cell damage, inflammatory cytokine production, and autoimmunity. MiRNA-204 is a highly enriched miRNA in human β-cells, and is released from dying β-cells into the serum. Serum miRNA-204 was increased in children with T1D, but not in those with type 2 diabetes, and was inversely correlated with cell functioning. Therefore, it could serve as a novel biomarker for T1D-associated β-cell damage [35].
Obesity-induced chronic low-grade inflammation leads to insulin resistance, type 2 diabetes mellitus, and metabolic syndrome [36,37]. MiRNAs play a regulatory role in multiple processes, including insulin signaling, adipokine expression, adipogenesis, lipid metabolism, and diet regulation. A high-fat maternal diet during pregnancy and lactation in mice resulted in altered hepatic miRNA-122 and miRNA-370 expression in the offspring [38]. Dysregulated miRNA-122 and miRNA-370 expression has been shown to modulate hepatic lipid metabolism and contribute to metabolic disturbances.In a severely obese pediatric cohort, miRNAs 34a, 122, and 192 were correlated with obesity-associated inflammatory markers, including tumor necrosis factor-α, interleukin-1 receptor antagonist, procalcitonin, and adiponectin [39]. In addition, the miRNA-122 concentration, which is correlated with homeostatic model assessment for insulin resistance and miRNA-192, was significantly elevated in obese participants. These miRNAs could serve as biomarkers for the unfavorable phenotype of childhood obesity, and may aid risk stratification for early intervention in childhood obesity.
Role of MiRNAs in cardiovascular disease
MiRNAs are also involved in cardiac development and function. Several miRNAs have been implicated in the development of the heart, and their dysregulation is associated with cardiovascular diseases, including congenital heart disease [40,41]. MiRNA-1 and miRNA-133 play a critical role in cardiac development, and miRNA-1 overexpression in the developing embryonic heart decreases the number of proliferating ventricular cardiomyocytes [42]. In mice, miRNA-1-2 deletion was lethal in approximately 50% of the embryos because of ventricular septal defects, while around 20% of the survivors had major cardiac defects [43]. MiRNA dysregulation has been reported in Down syndrome, the most common genetic cause of congenital heart defects. Overexpression in the heart of 5 human chromosome-21-derived miRNAs, including miRNA-99a, let-7c, miRNA-125b-2, miRNA-155, and miRNA-802, was also reported [44]. MiRNA-99a was associated with the suppression of cardiogenesis, let-7c was found to induce cardiogenesis, and miRNA-155 overexpression was found to inhibit necrosis.
MiRNAs are also important in the pathogenesis, treatment, and prognosis of Kawasaki disease (KD), which is the primary cause of acquired heart disease among children in developed countries [45]. MiRNA-223-3p overexpression has been reported in acute KD patients, in whom it may inhibit cytokines and alleviate vascular endothelial injury [46]. Zhang et al. [47] reported that lower miRNA-223 expression leads to severe coronary artery pathology in KD, and the presence of miRNA-223-3p may lead to the identification of patients at greatest risk for coronary artery pathology. As potential diagnostic biomarkers for KD, serum exosomal miRNA-1246, miRNA-4436b-5p, miRNA-197-3p, and miRNA-671-5p may be used to distinguish KD from other febrile conditions [48]. Furthermore, serum exosomal miRNA-328, miRNA-575, miRNA-134, and miRNA-671-5p could serve as biomarkers for KD and the outcomes of intravenous immunoglobulin therapy [49].
MiRNAs are also potential biomarkers for the diagnosis of heart failure and acute myocarditis [50-52]. The diagnosis of myocarditis remains a challenge. Blanco-Domínguez et al. [51] reported that mmu-miRNA-721 is synthesized by Th17 cells, and is present in the plasma of mice with acute autoimmune or viral myocarditis, but not in those with acute myocardial infarction. They reported that a novel human noncoding RNA (hsa-Chr8:96) with a similar sequence is associated with myocarditis, even after adjustment for age, sex, ejection fraction, and serum troponin levels. The area under the receiver operating characteristic curve for hsa-Chr8:96 to differentiate acute myocarditis and myocardial infarction was 0.927 (95% confidence interval, 0.879–0.975), with a sensitivity and specificity of >90%. Therefore, miRNAs could serve as alternative noninvasive biomarkers for the diagnosis and prognosis of cardiovascular diseases in the pediatric population.
MiRNAs in asthma
Asthma, which is the most common chronic illness among children, has a complex etiology involving genetic susceptibility, host factors, and environmental exposure [53]. Asthma is related to airway inflammation, tone control, and responsiveness, but the exact mechanism is unknown [54]. Ibrahim et al. [55] investigated Annexin A1 (ANXA1), an important anti-inflammatory mediator, and miRNA-196a-2, a targeted ANXA1 gene, in asthmatic children and controls. Asthmatic children had an increased serum ANXA1 level and decreased miRNA-196a-2 expression, indicating their role in asthma etiology and potential as diagnostic biomarkers and therapeutic targets. Tiwari et al. [56] demonstrated differential miRNA expression in asthmatic children among the 4 seasons. In particular, miRNA-328-3p and let-7d-3p were significantly associated with seasonal asthma symptoms and allergies. In addition, miRNAs were associated with asthmatic airways, airway remodeling, and virus-induced exacerbations. Several studies have been conducted on miRNA dysregulation in asthma, and miRNAs have been proposed as biomarkers for asthma status, severity, and treatment response.
MiRNAs in hematologic cancers
MiRNA dysregulation is a hallmark of childhood cancer [57]. The identification of molecular markers of pediatric tumors is important to predict prognosis, and may lead to the development of novel therapeutic approaches. MiRNAs play an important role in hematopoiesis and leukemogenesis.
In 2022, Calin et al. [58] reported frequent 13q14 deletions, and the downregulation of miRNA-15 and miRNA-16, in chronic lymphocytic leukemia. This was the first study of miRNA dysregulation associated with human disease. In 2004, the same group mapped 186 miRNAs and found that over 50% of the miRNA genes were located in cancer-associated genomic regions or fragile sites [59]. In addition, they identified the specific miRNA profile for human B-cell chronic lymphocytic leukemia [60]. In 2007, Mi et al. [61] analyzed the miRNA expression profiles of adult and pediatric cancer patients, and used them to accurately distinguish between acute lymphocytic leukemia and acute myeloid leukemia. In 2009, Ju et al. [62] analyzed differential miRNAs expression between childhood B-cell precursor acute lymphocytic leukemia and normal marrow samples, and found that miRNA-222, miRNA-339, and miRNA-142-3p were overexpressed, while miRNA-451 and miRNA-373 were downregulated. The clinical importance of miRNA profiles has also been confirmed based on their association with specific acute lymphocytic leukemia subtypes, treatment resistance, and event-free survival. Some miRNAs can be used to monitor disease progression, predict drug sensitivity, and assess prognosis. The role of miRNAs has been extensively studied in acute myeloid leukemia, chronic myelogenous leukemia, and lymphomas. Therefore, miRNAs could serve as diagnostic and prognostic molecular markers, and treatment targets, in hematological cancers.
MiRNAs in epilepsy
Epilepsy is a common and severe neurologic disorder characterized by recurrent seizures, increased mortality rates, and decreased social participation and quality of life [16]. Although antiepileptic drugs may control seizures in many patients, about one-third of cases are drug-resistant. It is necessary to identify individuals at risk for epilepsy, and to develop biomarkers and targeted therapies. The first study on miRNAs in human epilepsy was conducted in 2010, and reported elevated hippocampal miRNA-146a levels in temporal lobe epilepsy patients [63]. Since then, several studies have demonstrated that miRNAs are important regulators of gene expression in epilepsy.
Mooney et al. [64] manually curated the "EpimiRBase" database to provide comprehensive and up-to-date information on studies related to miRNAs and epilepsy. This fully searchable database includes brain and blood miRNA profiling studies, as well as functional studies. Specific miRNAs in the cerebrospinal fluid and blood may act as useful biomarkers for different types of brain injury, including prolonged seizures and refractory seizures [7]. Research on miRNAs may improve our understanding of the causes, treatments, and diagnosis of epilepsy.
Limitations of MiRNA studies
MiRNA research has several limitations. First, there are several methods for evaluating miRNAs, the most common of which are oligonucleotide microarray (microchip) and quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR). However, there is no gold standard for measuring miRNA expression [65]. It is considered good practice to profile miRNAs by microarray followed by validation through qRT-PCR [66]. However, there are no standard guidelines for performing and reporting such validation studies. Second, there is a lack of information about the effects of other factors, such as age, sex, sample type, ethnicity, and medications, on miRNA levels. Third, inconsistent standards for sample size calculation to ensure sufficient power, and a lack of reproducibility of results across studies, has led to skepticism regarding clinical applicability. Finally, the cost for miRNA research is still significant. Nevertheless, this is an exciting and growing area of research that is still in its early stages and remains relatively under-explored.
Conclusions
In this review, we have discussed the current evidence regarding the role of miRNAs in pediatric diseases. Although the field of miRNA research has advanced significantly over the past few years, it is still a promising area, especially in relation to pediatrics. Further research on miRNA expression may allow identification of novel biomarkers for the diagnosis, progression, and prognosis of pediatric diseases. The identification and characterization of disease-specific miRNAs and their targets may improve our understanding of the pathophysiology of these diseases at the molecular level and advance personalized pediatric medicine.
Notes
Conflicts of interest
No potential conflict of interest relevant to this article was reported.
Funding
This study was supported by the NRF grant, funded by the Korean government (MSIT; no. 2022R1G1A10 09727).