Characterization of gut microbiota in very low birth weight infants with versus without bronchopulmonary dysplasia
Article information
Abstract
Background
Gut-lung crosstalk is a pathway involving interactions between the gastrointestinal, respiratory, and immune systems. The immune responses of the gut and lungs are intricately linked, and previous studies demonstrated that the gut microbiota can influence systemic immune responses in the respiratory system as well as bronchopulmonary dysplasia (BPD).
Purpose
To analyze the composition of the gut microbiota in very low birth weight infants with versus without BPD.
Methods
Secondary data from a previous randomized controlled trial were analyzed. Microbiomes were analyzed using QIIME 2 software. Gut microbiota diversity and abundance were compared between groups.
Results
Fifty-one neonates were classified into the BPD (n=24) and non-BPD (n=27) groups, between which no differences were noted in the alpha and beta diversities of the gut microbiota. In both groups, Proteobacteria, Gammaproteobacteria, and Klebsiella were the predominant phylum, class, and genus in gut microbiota, respectively. Enterococcus, Acinetobacter, Elizabethkingia, Clostridium sensu stricto 1, Bacteroides, Streptococcus, and Serratia were more abundant, whereas Klebsiella, Faecalibacterium, Escherichia-Shigella, Enterobacter, Bifidobacterium, Veillonella, Staphylococcus, and Enterobacteriaceae were less abundant in the BPD versus non-BPD group. Faecalibacterium, Roseburia, Clostridium, Eubacterium, and Coprococcus were significantly more abundant in the non-BPD versus BPD group.
Conclusion
The alpha and beta diversities of the gut microbiota did not differ significantly between the BPD and non-BPD groups. However, in terms of relative abundance, the presence of common respiratory pathogens was notable in the BPD group. Conversely, the non-BPD group had a significantly higher prevalence of anaerobic taxa known for their capacity to produce butyrate, a key component of postbiotics.
Key message
Question: Does the gut microbiota differ between very low birth weight (VLBW) infants with versus without bronchopulmonary dysplasia (BPD)?
Finding: Common respiratory pathogens were notably elevated in the BPD group, whereas anaerobic and butyrate-producing taxa, key components of postbiotics, were dominant in the non-BPD group.
Meaning: In gut-lung communication, the interplay between the intestinal and respiratory systems may implicate pro- and postbiotics in VLBW infants with BPD.
Graphical abstract. NEC, necrotizing enterocolitis.
Introduction
Bronchopulmonary dysplasia (BPD) is a common long-term morbidity among infants with very low birth weight (VLBW; BW less than 1.5 kg) and very preterm (gestational age [GA] less than 32 weeks) infants. Preterm infants with BPD are more likely to die during early childhood or survive with severe developmental disabilities, which increases family stress and healthcare costs [1]. Longitudinal studies investigating the comprehensive and multidisciplinary care for premature infants have contributed to increasing survival rates without nosocomial infections, necrotizing enterocolitis (NEC), severe intraventricular hemorrhage, or severe retinopathy of prematurity [2,3]. However, reports on VLBW infants with BPD remained largely stagnant from 2008 to 2017 [2].
The Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network developed a BPD Outcome Estimator in 2022. The covariates in this tool were GA, body weight (BW), sex, antenatal steroids, highest mode of respiratory support and maximum fraction of inspired oxygen on the day of interest, and surgical NEC during postnatal days (PNA) 14 and 28 [1]. In addition to prevention of premature birth and the best respiratory support, nonrespiratory care, especially enteral nutrition or gut microbiome, may be associated with BPD in VLBW infants.
The gut microbiota of preterm infants is influenced by perinatal (pregnancy complications and mode of delivery), physiological (ethnicity, sex, growth, GA, and PNA), pharmacological (antibiotics or other medications), and genetic factors, as well as diet (milk, fortifiers, prebiotics, and probiotics) and neonatal intensive care unit (NICU) environment [4-7]. Microbes, their genes, and their metabolites function and interact optimally in the gut, brain, respiratory, and immune systems [8-10]. The gut-lung crosstalk is characterized by the interactions between the gut microbiota (specific gastrointestinal taxa, structural ligands, and secreted metabolites) and host responses [11,12]. The immune responses of the gut and lungs are intricately linked [11], and previous studies have demonstrated the potential of the gut microbiota to influence systemic immune responses in the respiratory system [11-13], as well as BPD in VLBW infants [14].
Some studies have investigated and reviewed the association between gut microbiota and neonatal BPD [14-17]. The primary aim of this study was to evaluate differences in the gut microbiota between VLBW infants with and without BPD.
Methods
1. Study design and participants
This case-control study analyzed secondary data on breast milk and sterile water intake in VLBW infants collected for a previous randomized controlled trial [18]. This study was conducted in the level IV NICU of the Songklanagarind Hospital, a university-affiliated hospital at Prince of Songkla University, between July 2018 and June 2020. The study was approved by the Ethics Committee of the Faculty of Medicine, Prince of Songkla University (REC. 60–455–01–1), and prospectively registered in the Thai Clinical Trials Registry (TCTR20180306002). Informed parental consent was obtained prior to enrollment.
The previous randomized trial [18] enrolled only inborn VLBW infants. Patients were excluded for the following reasons: maternal mortality, maternal or neonatal contraindications to breast milk, neonates considered to be nonviable or moribund by the attending neonatologist, neonates with prenatal or postnatal chromosomal syndromic anomalies, neonates with gut anomalies, or neonates whose parents did not provide consent to participate. To enhance the refinement of sequencing data for microbiota analysis, we evaluated rarefaction curves and established a sampling depth of 10,000 sequences for further analysis.
2. Feeding care
Gastric feeding was initiated within a few days of birth in stable neonates via an orogastric tube using mother’s milk, preterm formula, or both. Bovine milk-derived fortifier or concentrated formula milk were used for fortifications. Probiotics, prebiotics, and lactoferrin were not added during the study.
3. Fecal DNA collection and extraction
Fecal samples (0.5–1 g) were spontaneously collected by nurses directly from the diapers of each infant at 28 days of age into 1.5-mL microcentrifuge tubes using a sterile spatula. Specimens were frozen and stored at -20°C until later DNA extraction. DNA extraction was performed using the Zymobiomics (QIAamp Fast DNA Mini Kit; Qiagen) [19]. The 16S rRNA sequencing targeting the V3–V4 region was carried out at the Scientific Equipment Center, Prince of Songkla University, utilizing an Illumina MiSeq instrument (Illumina). QIIME 2 software package was used for quality filtering and demultiplexing the resulting sequencing data [20]. Taxonomic classification was based on the SILVA rRNA database.
4. Outcome and definitions
The primary outcome of this study was the composition of the gut microbiota in VLBW infants with and without BPD (BPD and non-BPD groups, respectively). BPD was defined as treatment with oxygen higher than 21% for at least 28 days [21].
Premature rupture of membranes was diagnosed more than 18 hours prior to delivery. Maternal antibiotics were defined as the history of maternal antibiotic use within 7 days before delivery. Patent ductus arteriosus was diagnosed by clinical and echocardiographic criteria. NEC was defined as stage II–III, diagnosed according to Bell’s criteria by clinical and radiographic evidences [22]. Culture-proven late-onset sepsis was defined as the fulfillment of clinical sepsis after 72 hours of life and bacterial or fungal growth in at least one blood culture of gram-negative bacteria or candidemia or at least 2 blood cultures of commensal organisms. Ventilator-associated pneumonia and pneumonia were diagnosed as the Centers for Disease Control and Prevention and National Healthcare Safety Network guidelines for infants <1 year old [23] by both a pediatrician and pediatric radiologist.
Oral care with mother’s own milk was calculated by the proportion of participants randomly receiving oral care via maternal breast milk divided by the total participants who received oral care—either mother’s own milk or sterile water [18]. The percentage volume for enteral breast milk was calculated by the volume of breast milk divided by the sum volume of breast milk and formula milk via gastric or oral feeding. The duration of antibiotic use was defined by the cumulative duration of antibiotic exposure until fecal sample collection. Time to full enteral feeds was defined as the time it takes for the VLBW neonates to tolerate volume of enteral feeds 120 mL/kg/day.
5. Statistical analysis
R software (ver. 4.3.2; The R Foundation for Statistical Computing, Vienna, Austria) was applied for comparison of baseline characteristics between neonates with and without BPD. Microbiome bioinformatic analysis was performed using QIIME 2 2021.11 [20].
Alpha- and beta-diversity metrics were calculated using the “diversity” plugin in QIIME 2. Alpha diversity metrics, reflecting species richness and evenness within bacterial populations, were estimated using Shannon’s Diversity Index, observed using operational taxonomic units, Pielou’s evenness, and Faith’s phylogenetic diversity. For alpha diversity, the P value was adjusted by variables with P values less than 0.2 from baseline characteristics (Table 1).
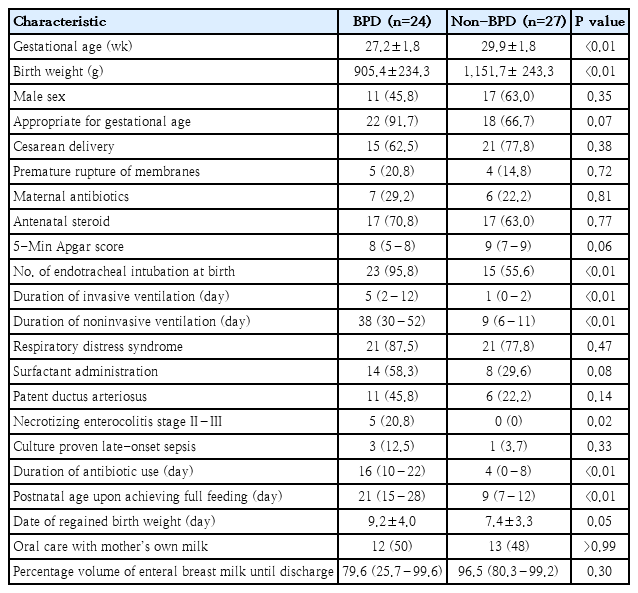
Baseline characteristics of neonates with versus without bronchopulmonary dysplasia (BPD and non-BPD groups)
Beta diversity metrics were estimated using unweighted and weighted UniFrac distance and represented as the q values (false discovery rate-corrected P value). For multivariate statistical methods, principal coordinate analysis determines the statistical significance of the UniFrac distance between 2 samples using a random (Monte Carlo) permutation test. In a permutational multivariate analysis of variance comparing the pseudo-F test result to that gained from random permutations of objects between the groups.
Relative and differential abundances of taxa were represented by percentage and linear discriminant analysis (LDA) effect size (LEfSe) [24], respectively. In the relative abundances of taxa, the main gut microbiota was defined as the percentage of taxa greater than 1% in either the BPD or non-BPD groups. The LEfSe method was applied to evaluate the significance of differences in operational taxonomic units between neonates with and without BPD. The threshold of LEfSe analysis for a discriminatory feature of the LDA score [log10] was set at 4.0.
Results
The randomized trial assessed 63 infants for eligibility, of whom 12 were excluded due to rarefaction curves and sampling depth <10,000 sequences. Finally, data from 51 neonates were analyzed for clinical factors and gut microbiota. The mean GA and BW was 28.6±2.2 weeks and 1,035.8±2,67.3 g. The median (interquartile range) 5-min Apgar score was 8 (7–9). Of the neonates, 28 (55%) were male, 40 (78%) were appropriate for GA, 36 (71%) were delivered by cesarean section, and 38 (75%) were endotracheal intubated.
The baseline characteristics of the BPD (47%, 24 of 51) and non-BPD (53%, 27 of 51) groups are shown in Table 1. VLBW infants with BPD had lower GA and BW but higher instances of endotracheal intubation at birth and NEC stage II–III, and longer duration of invasive and noninvasive ventilation, antibiotic use, time to full enteral feeding, and time to regained BW than did those without BPD. There were no cases of pneumonia and ventilator-associated pneumonia in either group.
Univariable analysis showed that the alpha and beta diversities of the gut microbiota did not differ between neonates with and without BPD (Table 2). Multivariable analysis showed that the alpha diversity was different between the groups, though not significantly, after adjusting for GA, BW, 5-min Apgar score, the proportion of patients appropriate for GA, endotracheal intubation at birth, surfactant administration, patent ductus arteriosus and NEC stage II–III, and duration of invasive and noninvasive ventilation, antibiotic use, time to full enteral feeding, and regained BW. Beta diversity was different between the groups, although not significantly, after false discovery rate correction, as shown in Table 2 and Fig. 1.
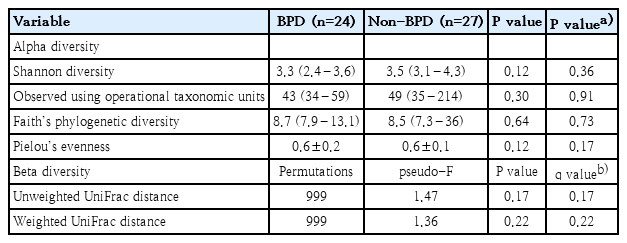
Gut microbiota biodiversity in preterm neonates with versus without bronchopulmonary dysplasia (BPD)
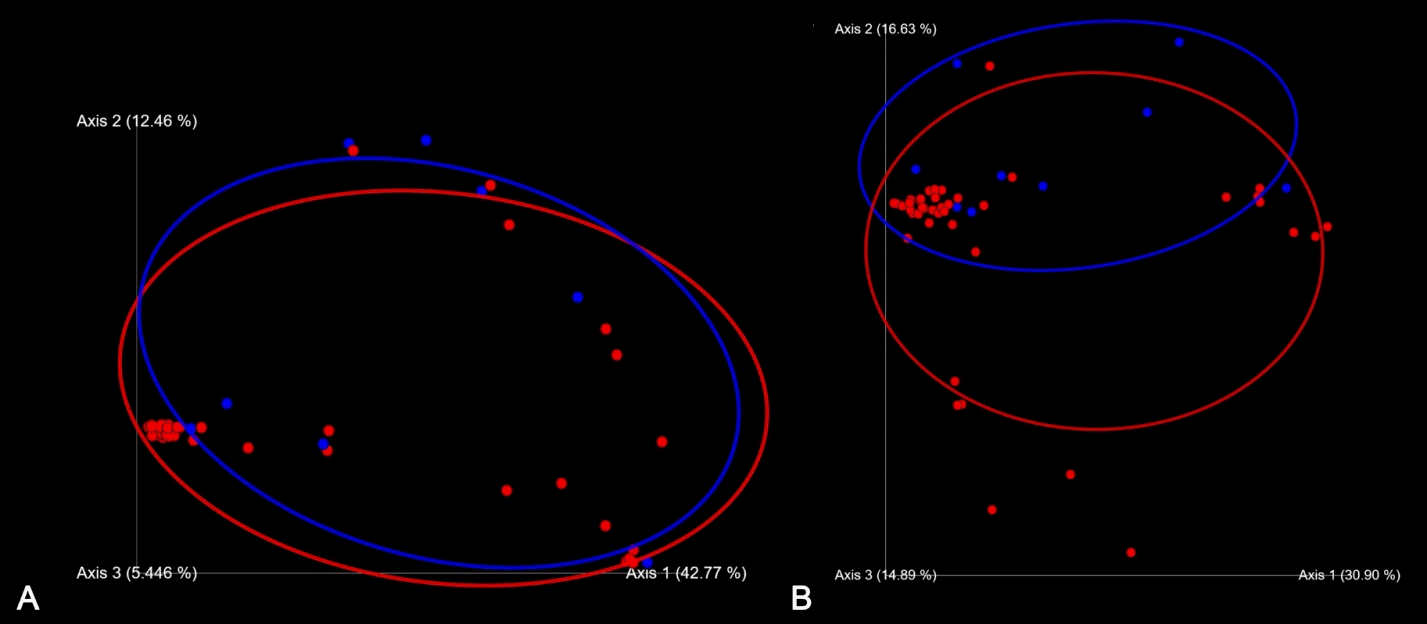
Beta diversity in very low birth weight infants with (blue dots) versus without (red dots) bronchopulmonary dysplasia. (A) Unweighted UniFrac distance. (B) Weighted UniFrac distance.
In the relative abundance of taxa, the main gut microbiota (>1%) between VLBW neonates with and without BPD is presented in Table 3. The highest percentage in phylum, class, and genus of gut microbiota in both the BPD and non-BPD groups was Proteobacteria, Gammaproteobacteria, and Klebsiella, respectively. Of the main gut bacteria, the BPD group had higher percentages of Enterococcus, Acinetobacter, Elizabethkingia, Clostridium sensu stricto 1, Bacteroides, Streptococcus, and Serratia; and lower percentages of Klebsiella, Faecalibacterium, Escherichia-Shigella, Enterobacter, Bifidobacterium, Veillonella, Staphylococcus, and unclassified Enterobacteriaceae than did the non-BPD group. Elizabethkingia, Stenotrophomonas, Corynebacterium, and Proteiniclasticum were found only in the BPD group, whereas Erysipelatoclostridium, UCG-002, Peptoniphilus, and Dialister were found only in the non-BPD group. Pseudomonas was found in the gut microbiota in 0.87% and 0.35% of patients in the BPD and non-BPD groups, respectively. A summary of the top 30 genera is presented in Fig. 2.
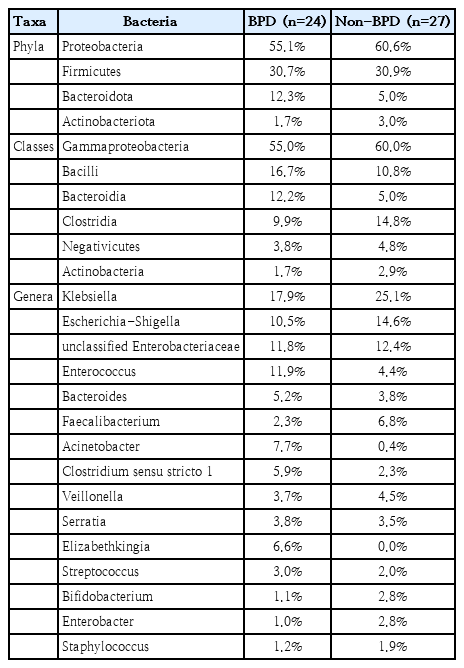
Relative abundances of the top taxa in the gut microbiota of preterm neonates with versus without bronchopulmonary dysplasia (BPD)
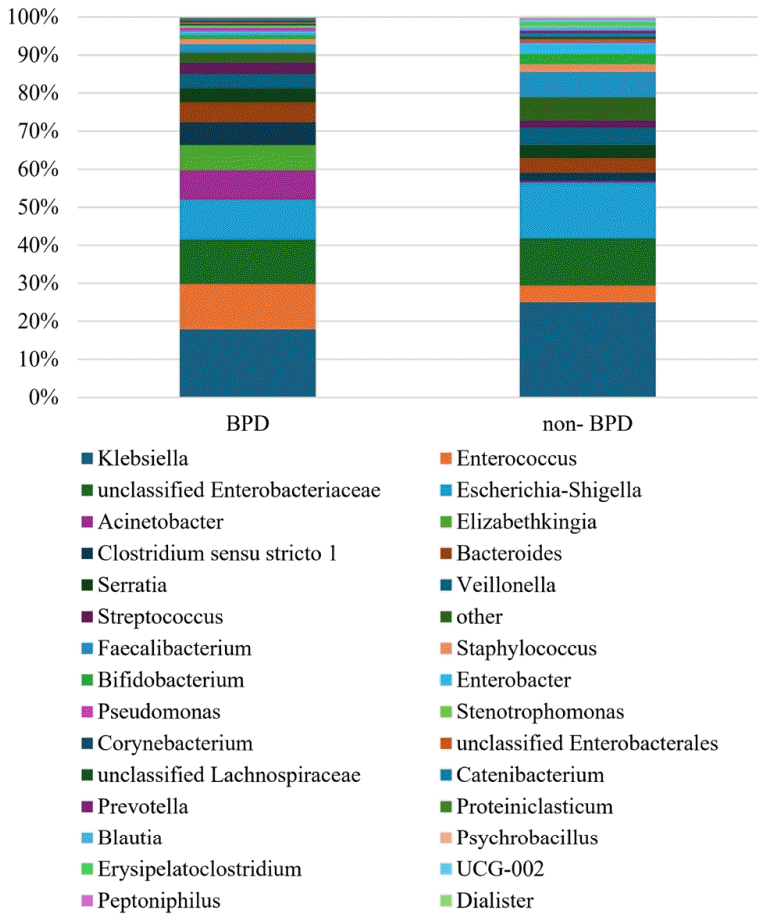
Relative abundance of the top 30 genera isolated from fecal samples of very low birth weight infants with versus without bronchopulmonary dysplasia (BPD).
In the differential abundance of taxa, LEfSe analysis with an LDA score >4 identified 25 key taxa that were significantly more abundant in neonates without BPD (Fig. 3A and B): genus Faecalibacterium, class Erysipelotrichi, order Erysipelotrichales, genus Catenibacterium, genus Roseburia, family Paraprevotellaceae, genus Prevotella, genus Clostridium, order Bacillales, genus Eubacterium, genus Ruminococcus, genus Rothia, family Odoribacteraceae, genus Lactonifactor, genus Dorea, genus Levilinea, genus Gemmiger, order SHA-20, genus Odoribacter, genus Coprococcus, family Rhodobacteraceae, genus Paraprevotella, genus Sporobacter, genus Pediococcus, genus Psychrobacillus. No taxa were significantly more abundant in neonates with BPD than in those without BPD.

Linear discriminant analysis effect size showing differentially abundant microbial clades in the fecal samples. (A) Red bar chart represents the key to significantly abundant taxa in neonates without bronchopulmonary dysplasia (non- BPD). (B) Cladogram showing significantly abundant clades (red circles represent the non-BPD group, whereas red bars [a–g] do not include the genus and species levels). After the exclusion of neonates with necrotizing enterocolitis. (C) The significantly abundant taxa in the non-BPD group represented as a red bar chart. (D) Cladogram showing significantly abundant clades (red circles represent the non-BPD group, while red bars [a] do not include genus and species levels). (A–D) Neonates with BPD did not show a significant abundance of taxa (B and D): Nonsignificant microbial clades (dark yellow circles). LDA, linear discriminant analysis.
Following the exclusion of VLBW neonates with NEC (n=5), LEfSe analysis with an LDA score >4 identified 5 key taxa that were significantly more abundant in neonates without BPD without NEC (Fig. 3C and D): family Paraprevotellaceae, genus Rothia, genus Roseburia, genus Clostridium, and genus Psychrobacillus. When neonates with NEC were excluded, no taxa were significantly more abundant in the BPD group than in the non-BPD group.
Discussion
The clinical factors found to be associated with BPD in the present study largely matched those of similar previous studies and additionally include earlier preterm births, intubation, NEC, and longer duration of ventilatory support, and antibiotic use. Lower GA and BW preterm [1,14-16], longer duration of invasive ventilation [15,16], and antibiotic use [16] are well-documented risk factors for BPD, and surgical NEC has been associated with BPD [1]. VLBW infants with NEC were only found in the BPD group. Previous studies have demonstrated the differences in the gut microbiota between VLBW neonates with BPD and those without BPD, and the baseline characteristics were compared with those in this study [14-16]. The mean GA (27 weeks) and BW (0.9 kg) in this study were similar to those reported in 2 previous studies [15,16] but higher than those reported by Ryan et al. [14] (GA 25 weeks and BW 0.7 kg). The duration of invasive ventilation in this study was similar to that reported by Zhang et al. [15] but shorter than that reported by Chen et al. [16] (23.5 days). All severity grades of BPD were observed in the present study and the study by Zhang et al. [15], whereas some prior studies only identified moderate-to-severe BPD [14,16].
In both the unadjusted and adjusted models, the alpha and beta diversities did not differ significantly between the BPD and non-BPD groups. Previously, no significant differences in alpha and beta diversities were reported in a mice study [25] between the BPD and control groups on day 7 and 21 (n=10), and only significant differences in alpha diversity were reported between the BPD and control groups in the first month in human preterm infants at <29 weeks GA (n=20) [14]. Conversely, alpha diversity indices on day 28 were reported to be significantly lower for preterm infants with BPD who were born at <32 weeks GA (n=8) [16] or <30 weeks GA (n=18) [15] than for preterm infants without BPD.
Most of the taxa in the gut microbiome differ across ages, from preterm-, full-term neonates, infants, children, and adults. In adults, the most abundant taxa have been reported to be from the Firmicutes and Bacteroidota phyla, whereas relatively few taxa from the Proteobacteria, Actinobacteria, and Fusobacteria phyla are present [26]. In children (3 years old), 99% of the gut microbiome is comprised of obligate anaerobes, including Firmicutes (Clostridium coccoides [17%] and Clostridium leptum [34%]), Bacteroidota (Bacteroides fragilis [21%]), and Actinobacteria (bifidobacteria [22%]) [27]. In infants (6 months old), the most abundant taxon is Actinobacteria (bifidobacteria [42%]), followed by Proteobacteria (enterobacteria [34%]) and Bacteroidota (B. fragilis group [19%]) [27]. In full-term and breast-fed neonates, Proteobacteria (Enterobacteriaceae) and Actinobacteria (Bifidobacteriaceae) account for more than 50% of the composition of the gut microbiota [27,28]. The gut microbiome in VLBW infants is influenced by several factors, including GA, BW, cesarean delivery, premature rupture of membranes, maternal and neonatal antibiotics, breastfeeding, and duration of NICU stay [29]. The 4 major phyla present in the fecal microbiota in VLBW [30] and very preterm infants [16] with BPD reported in previous studies (Proteobacteria [58.7% and 52.2%], Firmicutes [37.9% and 39.3%], Bacteroidetes [2.0% and 2.8%], and Actinobacteria [1.4% and 4.2%]) were similar to those found in this study. Although the number of articles published on this topic is limited [16,30], the gut microbiota in VLBW infants showed consistency with previous papers. We were able to conclude the whole picture from the results of our and prior studies, the dominant phyla in neonates (preterm [16,30] and full-term [27,28]), infants [27], and children (until adults) [26,27] are Proteobacteria, Actinobacteria, and Firmicutes, respectively.
In the differential abundance analysis, the predominant gut microbiota identified in the non-BPD group were butyrate-producing bacteria (Faecalibacterium, Roseburia, Clostridium, Eubacterium, and Coprococcus). Most butyrate producers belong to the Clostridium cluster (Clostridium IV or C. leptum group [such as Faecalibacterium and Ruminococcus] and Clostridium XIVa or C. coccoides group [such as Roseburia, Eubacterium, and Coprococcus]) of the phylum Firmicutes. Butyrate is used to generate energy for colonocytes and maintains gut barrier integrity by immune modulation and epigenetic regulation [31]. Colonic butyrate producers shape the gut microbial community by antimicrobial substances, and anti-inflammatory molecules [32]. Butyrate is a short-chain fatty acid produced by bacteria in the colon and is classified as a postbiotic (a byproduct of probiotics that confers health benefits). For example, Bifidobacterium produces acetate and lactate, which are further metabolized by butyrate producers to generate butyrate; this in turn supports the abundance of Bifidobacterium [33]. The interaction of postbiotics, probiotics, and butyrate producers in the gut microbiota show cross-feeding, positive synbiotic (combination of prebiotics, probiotics, and postbiotics) and symbiotic (between probiotics, such as Bifidobacterium, and butyrate producers, such as Faecalibacterium, Roseburia, Anaerostipes, and Eubacterium) relationships [32]. In the non-BPD group, the predominance of butyrate-producing bacteria in the gut microbiota may help to build the healthy intestinal environment, providing benefit to systemic organs, including the respiratory tract.
NEC has also been reported to be associated with BPD [1] and was found only in the BPD group in this study. After VLBW infants with NEC were excluded from the BPD group, the predominant gut taxa in the non-BPD group comprised both anaerobic (Paraprevotellaceae, Roseburia, and Clostridium) and aerobic bacteria (Rothia, and Psychrobacillus). Preterm infants with BPD are prone to long-term oxygen exposure (at least 28 days) [34], which may interfere with the anaerobic environment in the gut during the first month [16].
This study has some notable strengths. First, we demonstrated the differences in the gut microbiota in VLBW neonates with BPD (24 neonates) and those without BPD (27 neonates), in addition to previous studies (20 [14], 18 [15], and 8 [16] neonates with BPD). Second, the alpha diversity, beta diversity, and LEfSe were adjusted accordingly by clinical factors, false discovery rate correction, and the exclusion of neonates with NEC, respectively. Third, the results indicate the role of BPD and the intestinal microbiota regarding the gut-lung axis and their communication and community by pro-, post-, sym- and synbiotics in neonatal chronic lung diseases. A comprehensive understanding of the gut microbiota, particularly for postbiotics for microbiome-targeting therapies may provide multimodal strategies for BPD and allow for tailored early interventions and targeted treatments. However, this study has some limitations. First, the airway microbiome was not tested to confirm the association between gut microbiota and BPD. Second, the gut microbiomes may have been influenced by some confounding factors; however, the biodiversity and LEfSe analysis were adjusted by clinical factors, false discovery rate correction, and the exclusion of neonates with NEC to improve the internal validity of the study results. Third, data on the volume of breast milk was not collected in case breastfeeding affected the gut microbiome. Fourth, although a new definition of BPD was presented in 2019, this study used the classic definition, originally described in 2001, because the years of fecal collection in this randomized controlled trial (2018–2020) predated the introduction of the new BPD diagnosis (2019). Moreover, most published papers have used the previous definition of BPD. Fifth, we did not publish any subgroup analysis of the severity of BPD due to the limited number of participants. Finally, our study was performed on postnatal day 28, in accordance with previous studies [15,16]. Further studies are required to determine the long-term effects of BPD, including BPD according to the new definition presented in 2019, on the gut microbiome.
In conclusion, the biodiversity in the gut microbiome did not differ significantly between the BPD and non-BPD groups. However, the relative abundances of Proteobacteria, Actinobacteriota, and Firmicutes were lower, whereas that of Bacteroidota was higher, in the BPD group than in the non-BPD group. In the non-BPD group, anaerobic and butyrate-producing taxa were dominant in terms of their differential abundances. From the viewpoint of microbial therapeutics, leveraging postbiotics from these butyrate producers could be a possible preventive measure [32], as could treatment with predatory bacteria as living antibiotics [10,35]. In the context of gut-lung communication, the interplay between the intestinal and respiratory systems may implicate probiotics and postbiotics in VLBW infants with BPD.
Notes
Conflicts of interest
No potential conflict of interest relevant to this article was reported.
Funding
This study was funded by Targeted Research Grants from the Faculty of Medicine, Prince of Songkla University (REC. 60-455-01-1).
Acknowledgments
Clinical Trial Registration: This trial was prospectively registered at Thai Clinical Trials (https://www.thaiclinicaltrials.org/export/pdf/TCTR20180306002; first posted registration: March 6, 2018).
Author Contribution
Conceptualization: AT, KS, KS; Data curation: AT, MP, GM, SD; Formal analysis: AT, KS, KS; Funding acquisition: AT; Methodology: AT, MP; Project administration: AT, GM, SD; Visualization: AT; Writingoriginal draft: AT; Writing-review & editing: AT, MP, GM, SD, KS, KS

